R语言可视化学习笔记之基因组数据可视化
 本文主要利用ggpubr包来探索基因组数据,主要是可视化TCGA基因组数据的基因表达谱。
本文主要利用ggpubr包来探索基因组数据,主要是可视化TCGA基因组数据的基因表达谱。
library(ggpubr)#加载包
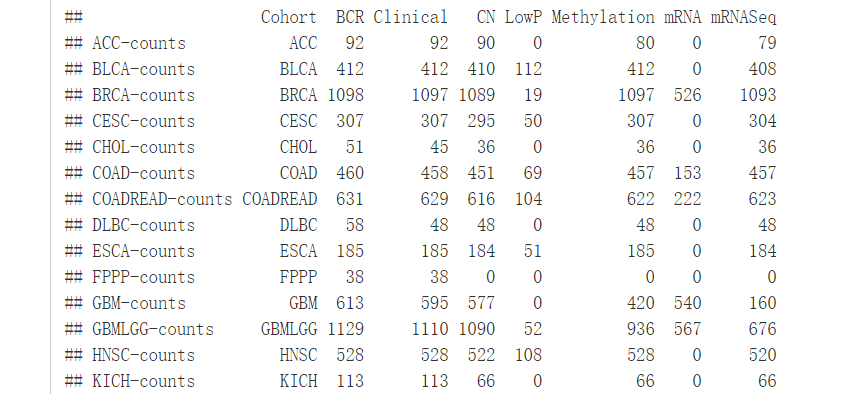
TCGA是一个包含大量癌症数据的数据库,由Marcin Kosinski创建的RTCGA包可以让我们很方便的获取这些数据。主要有三个包:RTCGA、RTCGA.clininal、RTCGA.mRNA。安装方法如下:
#Load the bioconductor installersource
("https://bioconductor.org/biocLite.R"")
#设置镜像,这里我们选择中科大的镜像options(BioC_mirror="https://ustc.edu.cn/bioc")
#下载包
biocLite("RTCGA")
biocLite("RTCGA.clininal")
biocLite("RTCGA.mRNA")
library(RTCGA)
#查看每一种癌症的数据集
infoTCGA()
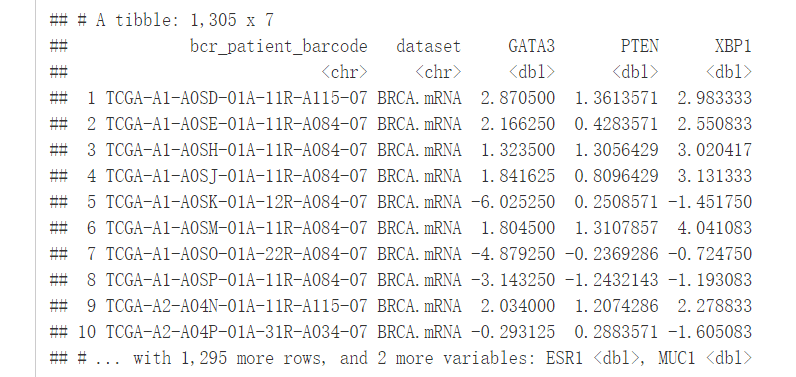
RTCGA包里的函数expressionTCGA()可以十分方便地从不同数据集中提取基因的表达值,下面我们将从三个数据集BRCA(乳腺癌)、OV(卵巢癌)、LUSC(肺癌)中提取五个基因的表达值。
library(RTCGA)
library(RTCGA.mRNA)
expr <- expressionsTCGA(BRCA.mRNA, OV.mRNA, LUSC.mRNA,
extract.cols = c("GATA3", "PTEN", "XBP1", "ESR1", "MUC1"))
expr
 查看每个数据集中的样品数量
查看每个数据集中的样品数量
nb_samples <- table(expr$dataset)nb_samples
##
## BRCA.mRNA LUSC.mRNA OV.mRNA
## 590 154 561
为了方便,我们将部分数据集名称简化
expr$dataset <- gsub(pattern = ".mRNA", replacement = "", expr$dataset)
expr$bcr_patient_barcode <- paste0(expr$dataset, c(1:590, 1:561, 1:154))
expr

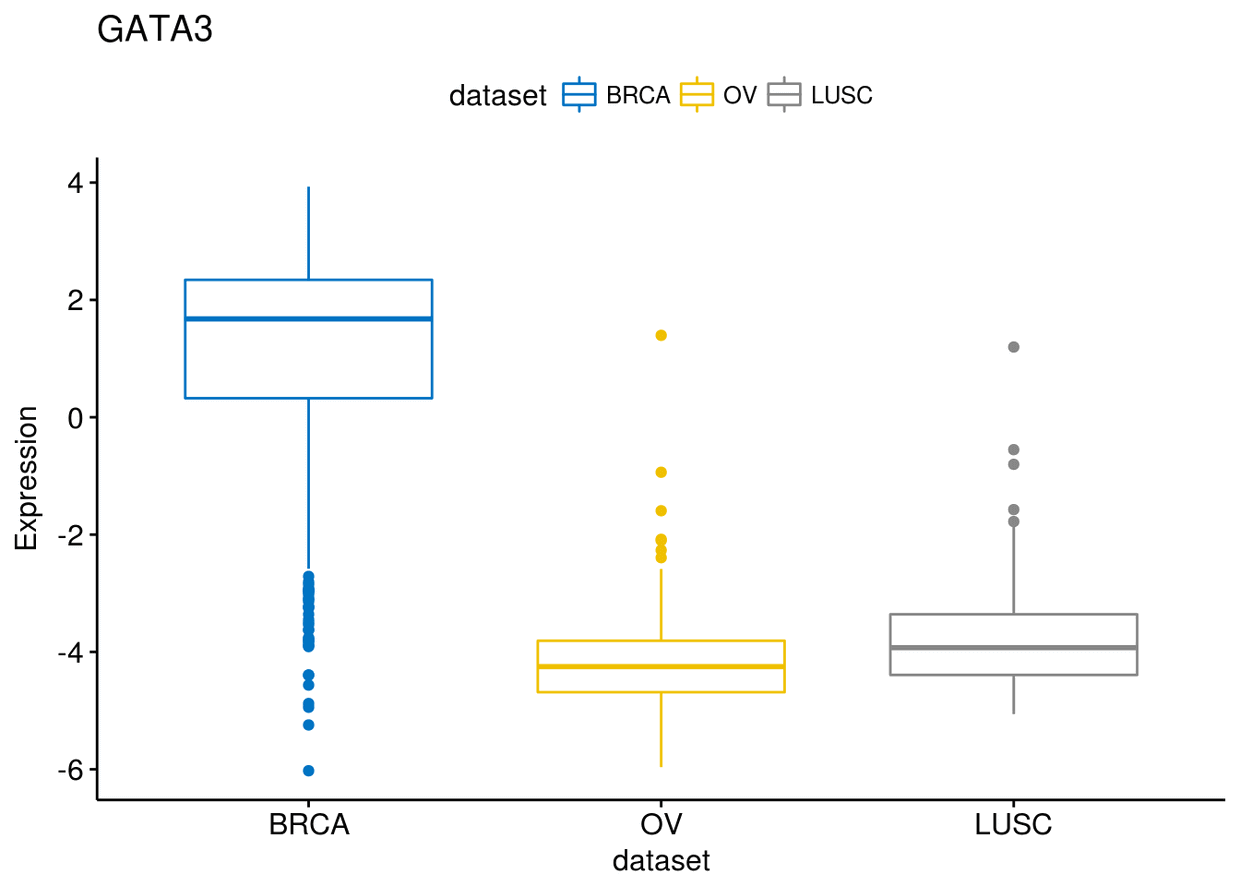
接下来绘制图形: 1、箱线图
library(ggpubr)
ggboxplot(expr, x="dataset", y="GATA3", title="GATA3", ylab = "Expression",
color = "dataset", palette = "jco")
 我们可以一次性绘制多个基因,然后一一查看,而不用每次写代码:
我们可以一次性绘制多个基因,然后一一查看,而不用每次写代码:
#Creat a list of plots
p <- ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1", "ESR1", "MUC1"),
title = c("GATA3", "PTEN", "XBP1", "ESR1", "MUC1"),
ylab = "EXpression", color = "dataset", palette = "jco")
#接下来一一查看每个plot
p$GATA3
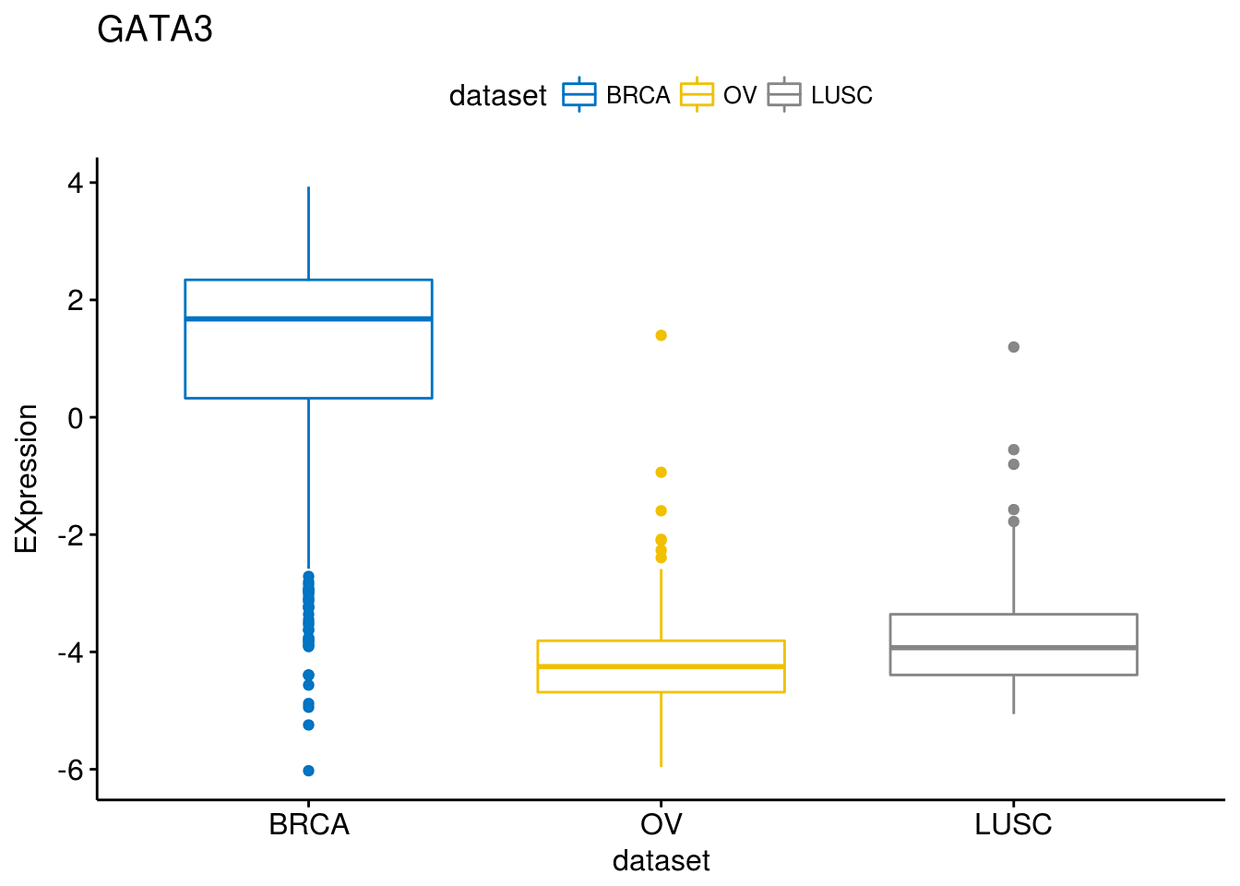
p$PTEN
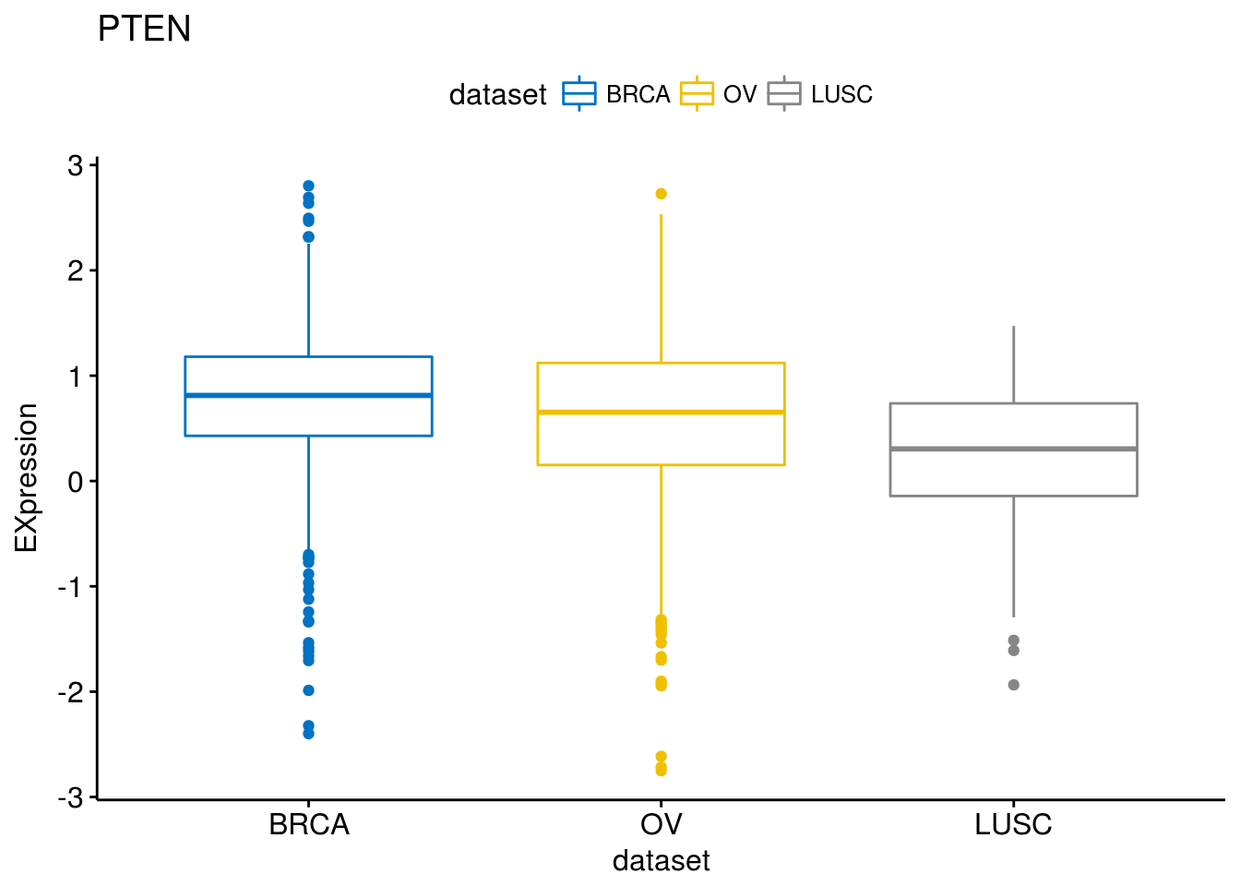
p$XBP1
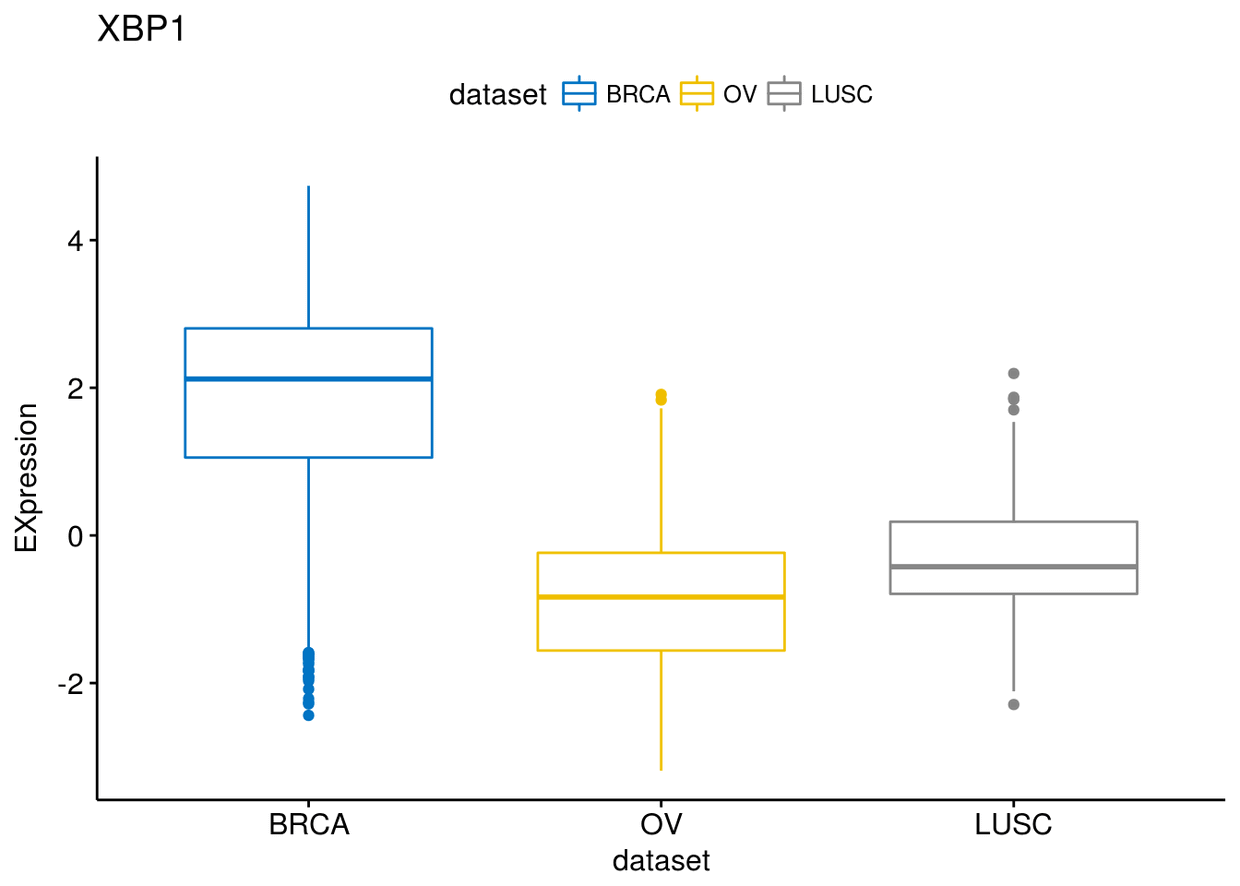
p$ESR1
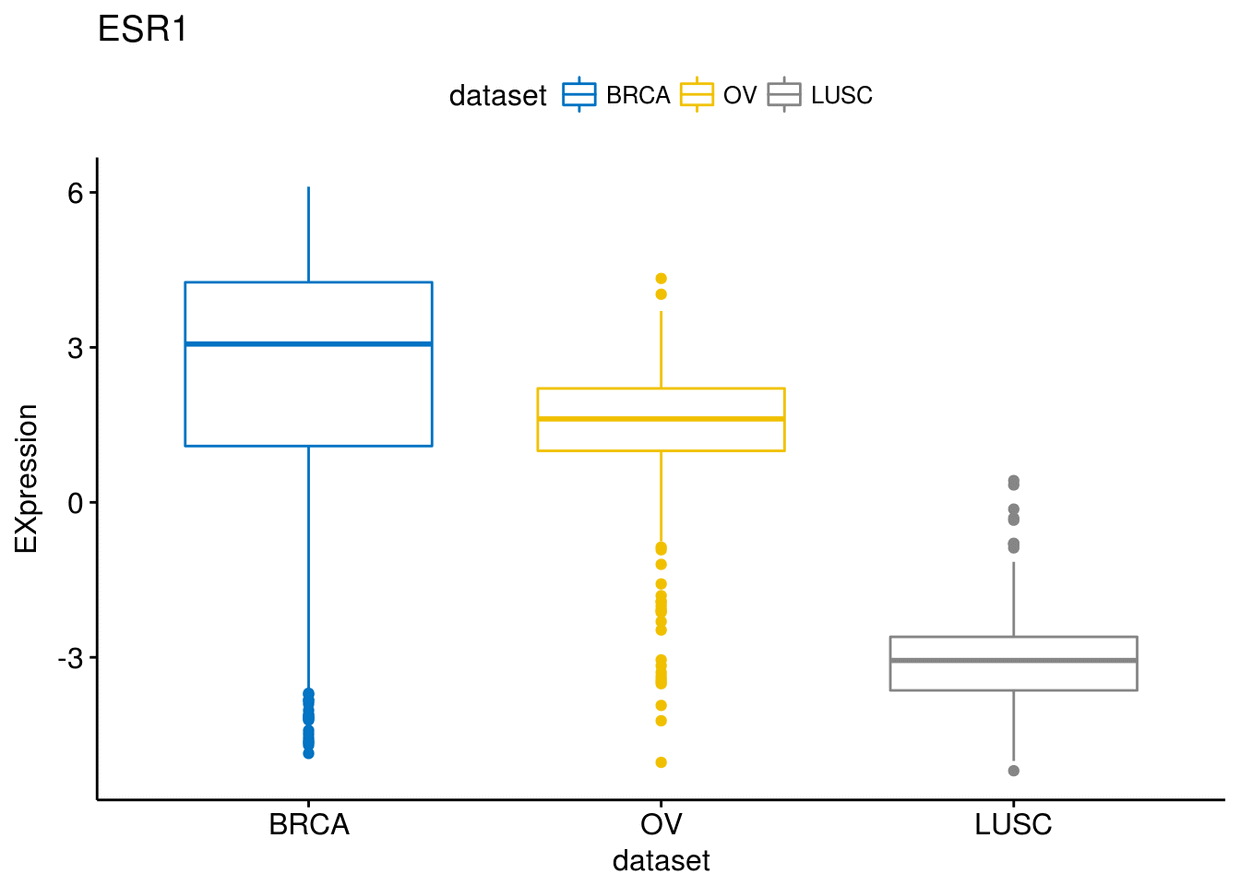
p$MUC1
 当一次性绘制多个基因时,xlab,ylab,title也可以是一个跟y等长的向量。 接下来就是添加p-value以及显著性了
当一次性绘制多个基因时,xlab,ylab,title也可以是一个跟y等长的向量。 接下来就是添加p-value以及显著性了
my_comparisons <- list(c("BRCA", "OV"), c("OV", "LUSC"))
ggboxplot(expr, x="dataset", y="GATA3", title = "GATA3", ylab = "Expression",
color = "dataset", palette = "jco")+ stat_compare_means(comparisons = my_comparisons)
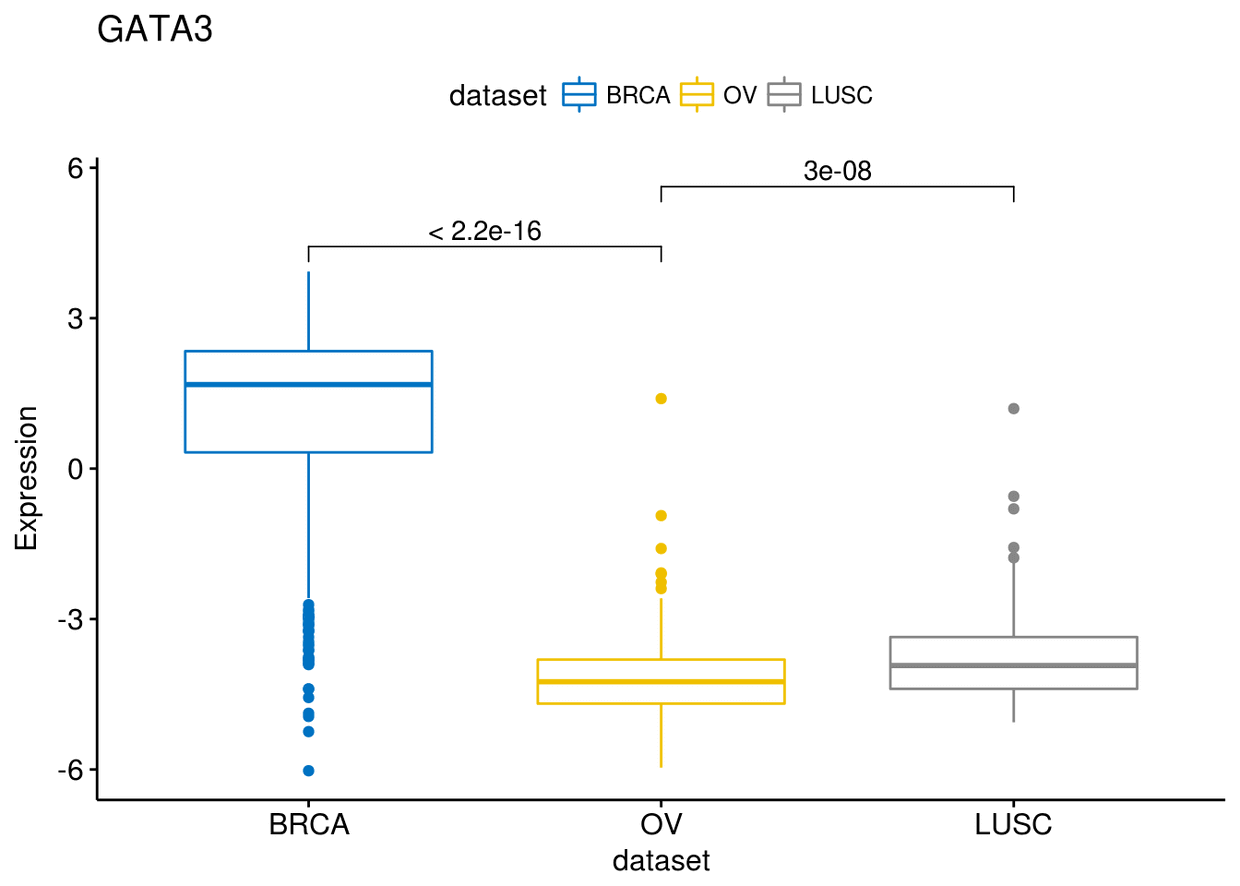 也可以查看每个类型中每一个基因的比较:
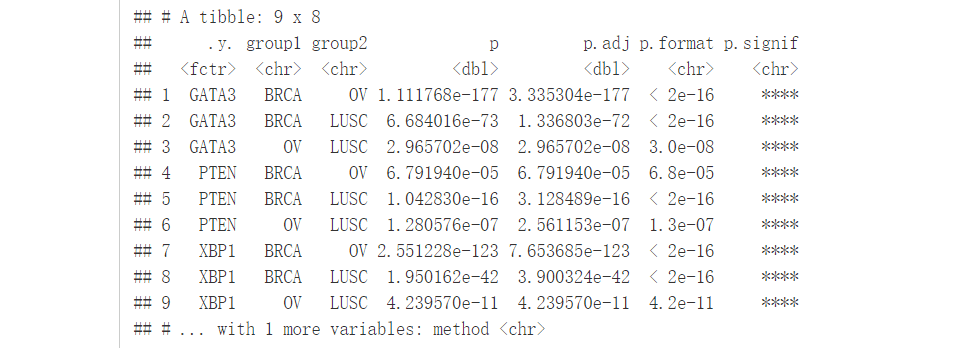
也可以查看每个类型中每一个基因的比较:
compare_means(c(GATA3, PTEN, XBP1)~dataset, data = expr)
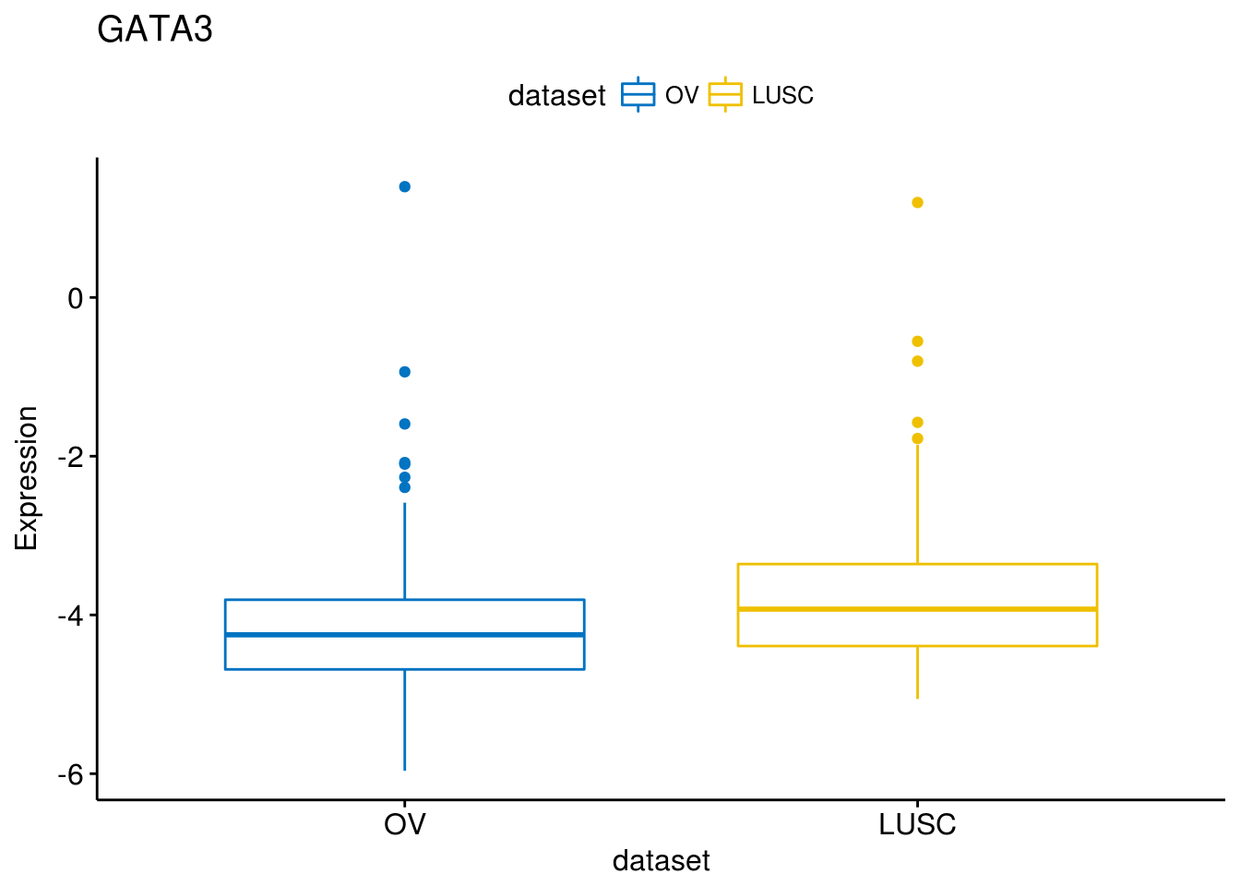
可以通过select以及remove来决定比较那几个类型,比如这里我们只比较BRCA和OV
ggboxplot(expr, x="dataset", y="GATA3", title = "GATA3", ylab = "Expression",
color = "dataset", palette = "jco", select = c("BRCA", "OV"))#通过select选择

ggboxplot(expr, x="dataset", y="GATA3", title = "GATA3", ylab = "Expression",
color = "dataset", palette = "jco", remove = "BRCA")#通过remove选择
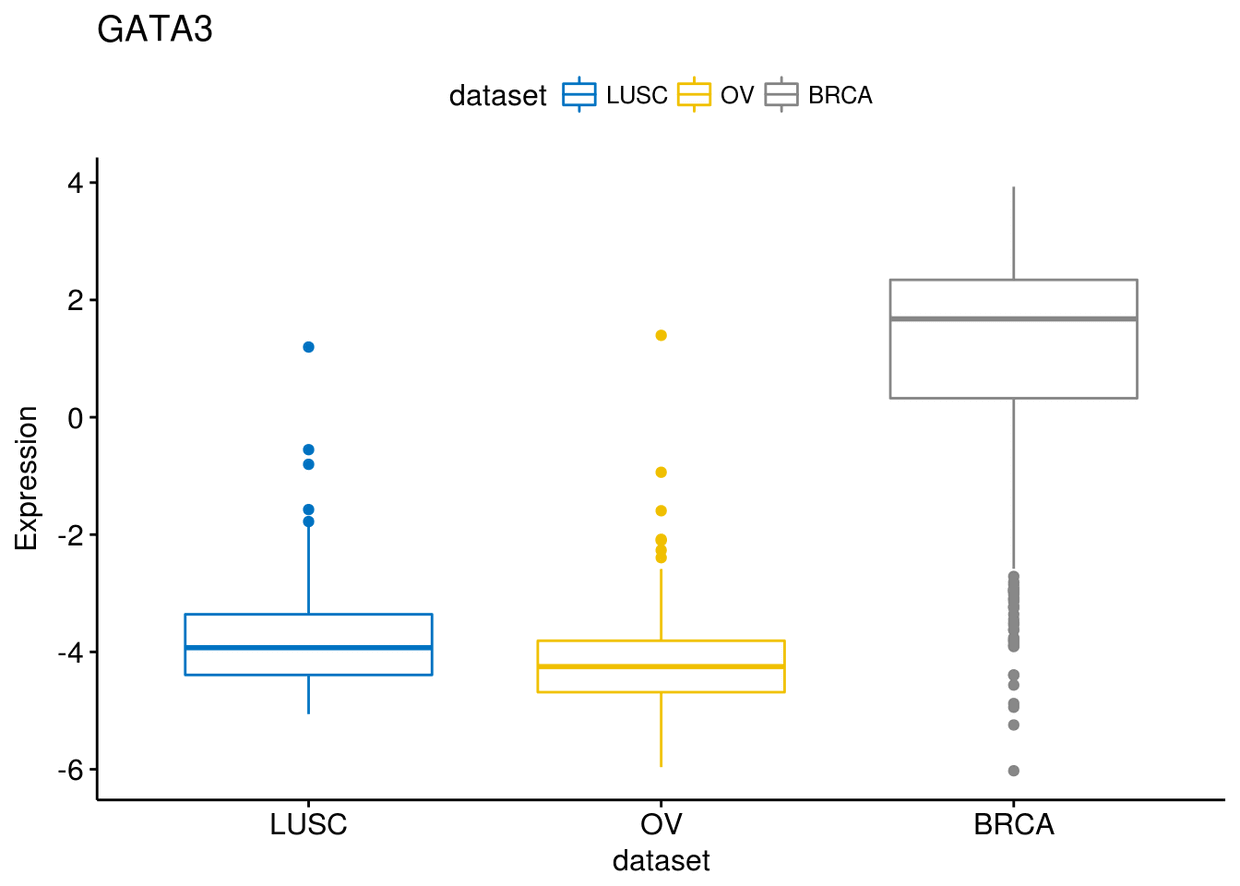
 通过order来改变各类型在x轴上的顺序
通过order来改变各类型在x轴上的顺序
ggboxplot(expr, x="dataset", y="GATA3", title = "GATA3", ylab = "Expression",
color = "dataset", palette = "jco", order = c("LUSC", "OV", "BRCA"))
 通过rotate=TRUE来变换坐标轴
通过rotate=TRUE来变换坐标轴
ggboxplot(expr, x="dataset", y="GATA3", title = "GATA3", ylab = "Expression",
color = "dataset", palette = "jco", rotate=TRUE)
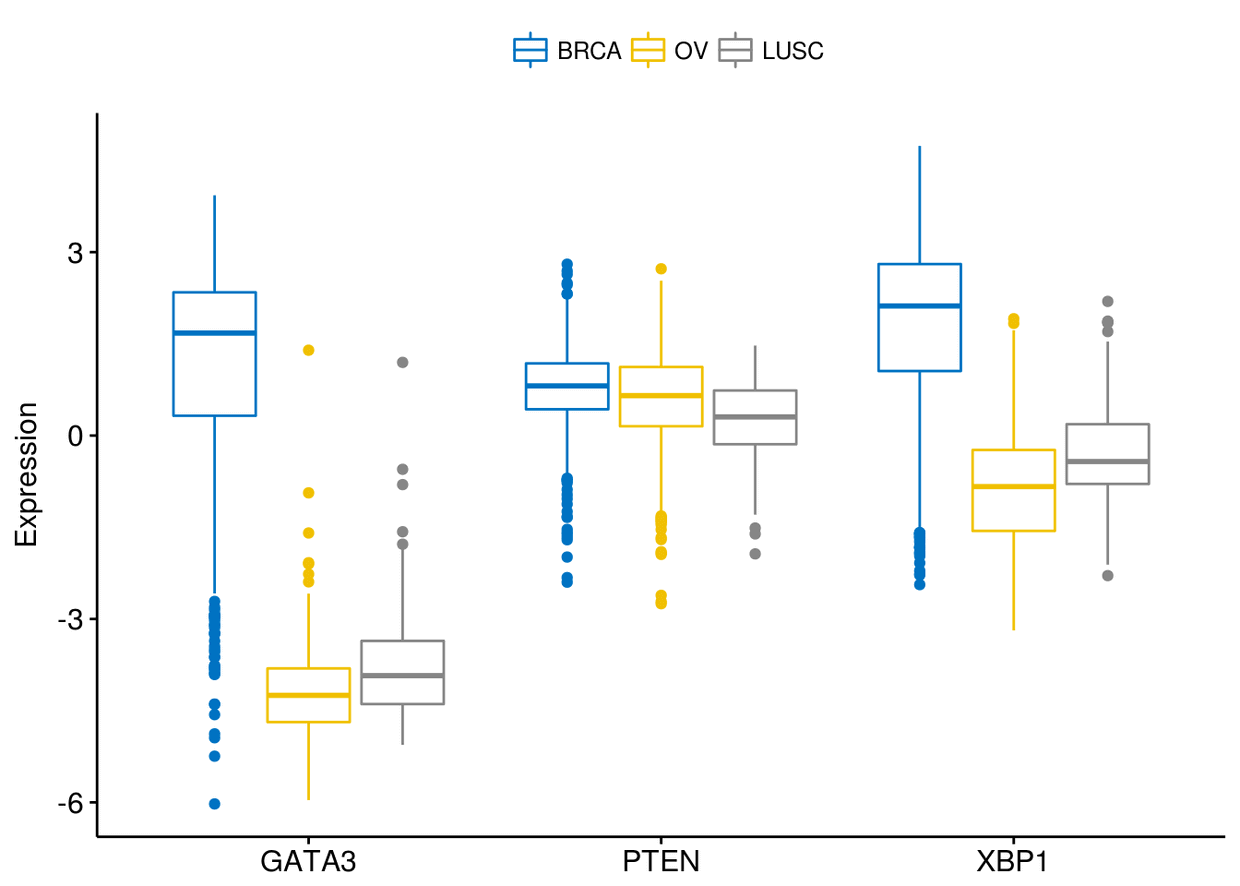
 通过combine=TRUE来进行分面(类似于facet)
通过combine=TRUE来进行分面(类似于facet)
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), ylab = "Expression",
color = "dataset", palette = "jco", combine = TRUE)
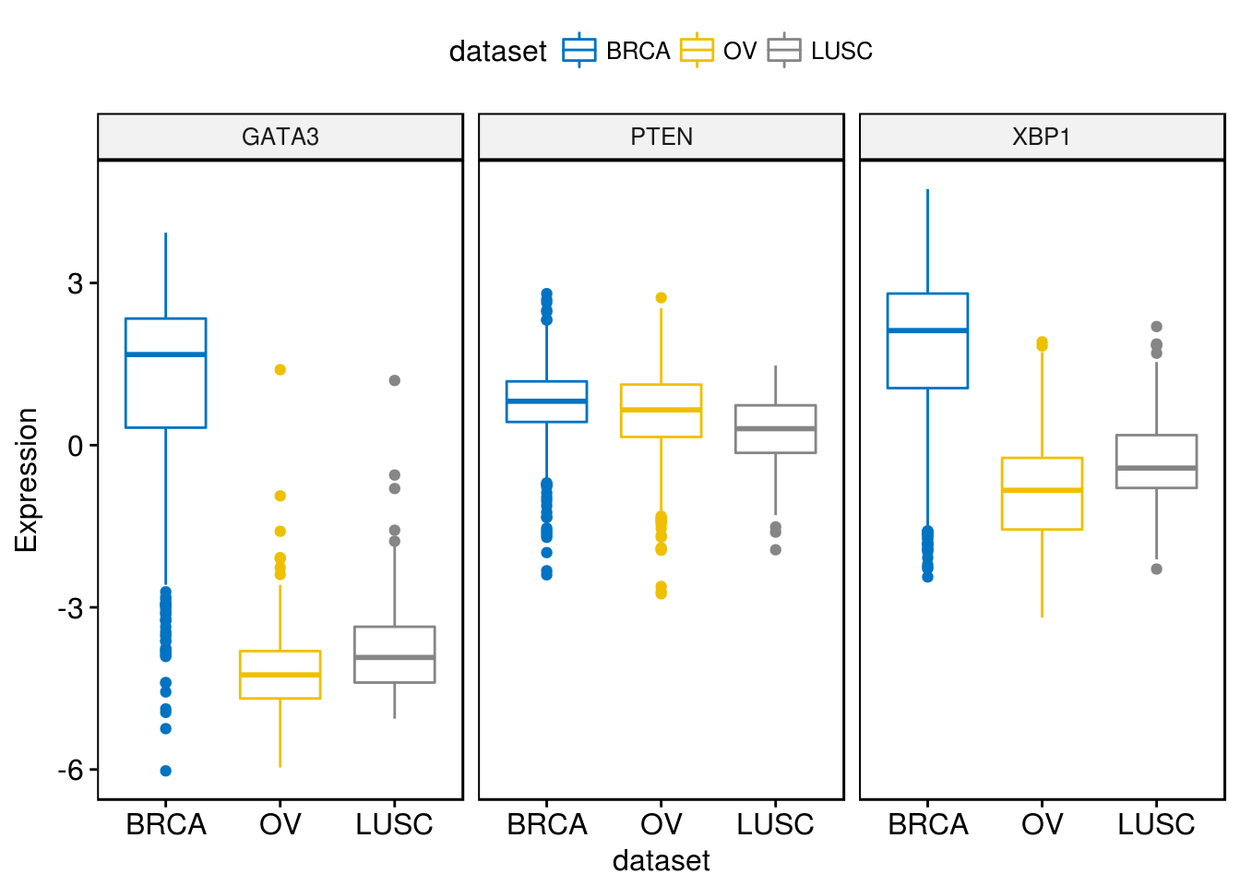 通过merge=TRUE或者merge=“axis”将三个类型的plot绘制在一个panel中
通过merge=TRUE或者merge=“axis”将三个类型的plot绘制在一个panel中
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), ylab = "Expression",
color = "dataset", palette = "jco", merge = TRUE)
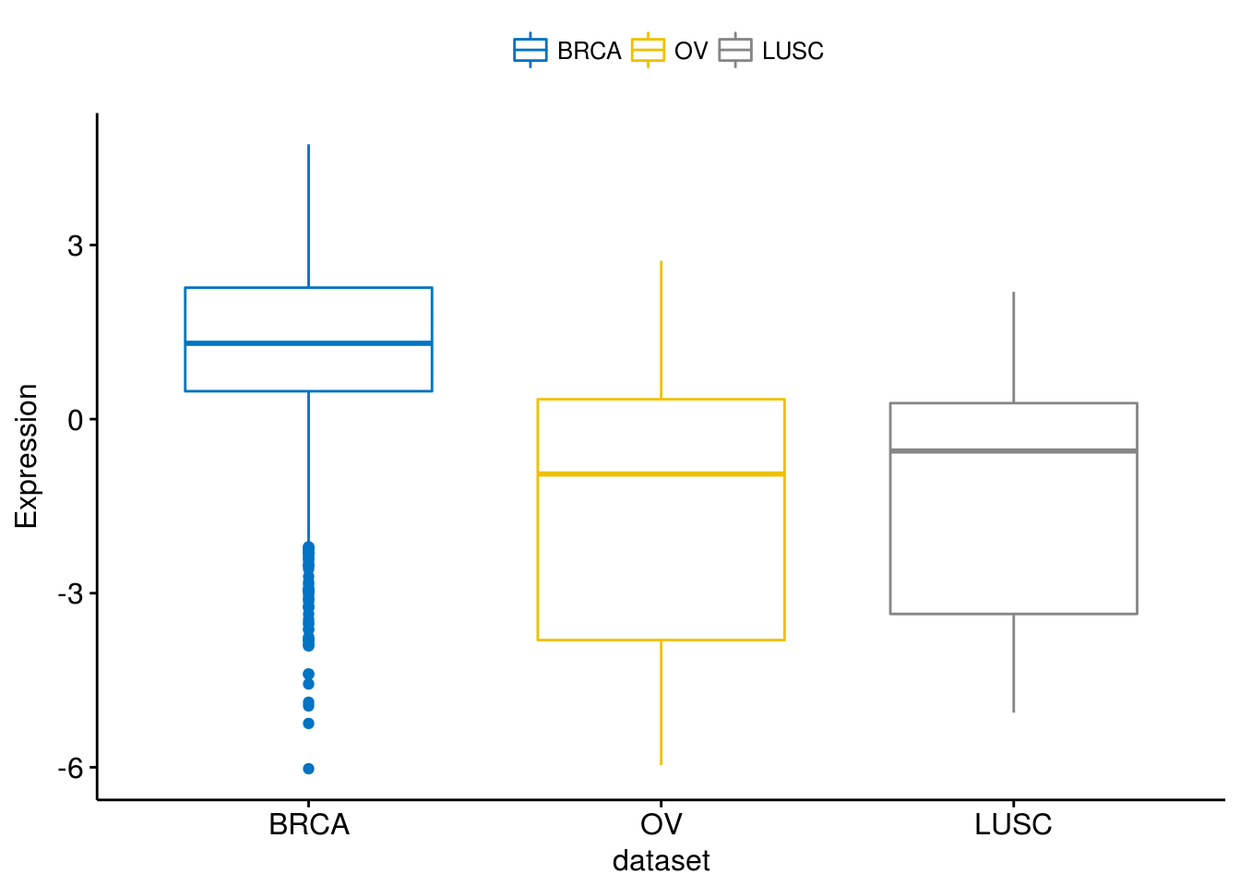 通过merge=flip利用不同癌症类型进行group
通过merge=flip利用不同癌症类型进行group
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"),
ylab = "Expression", palette = "jco", merge = "flip")
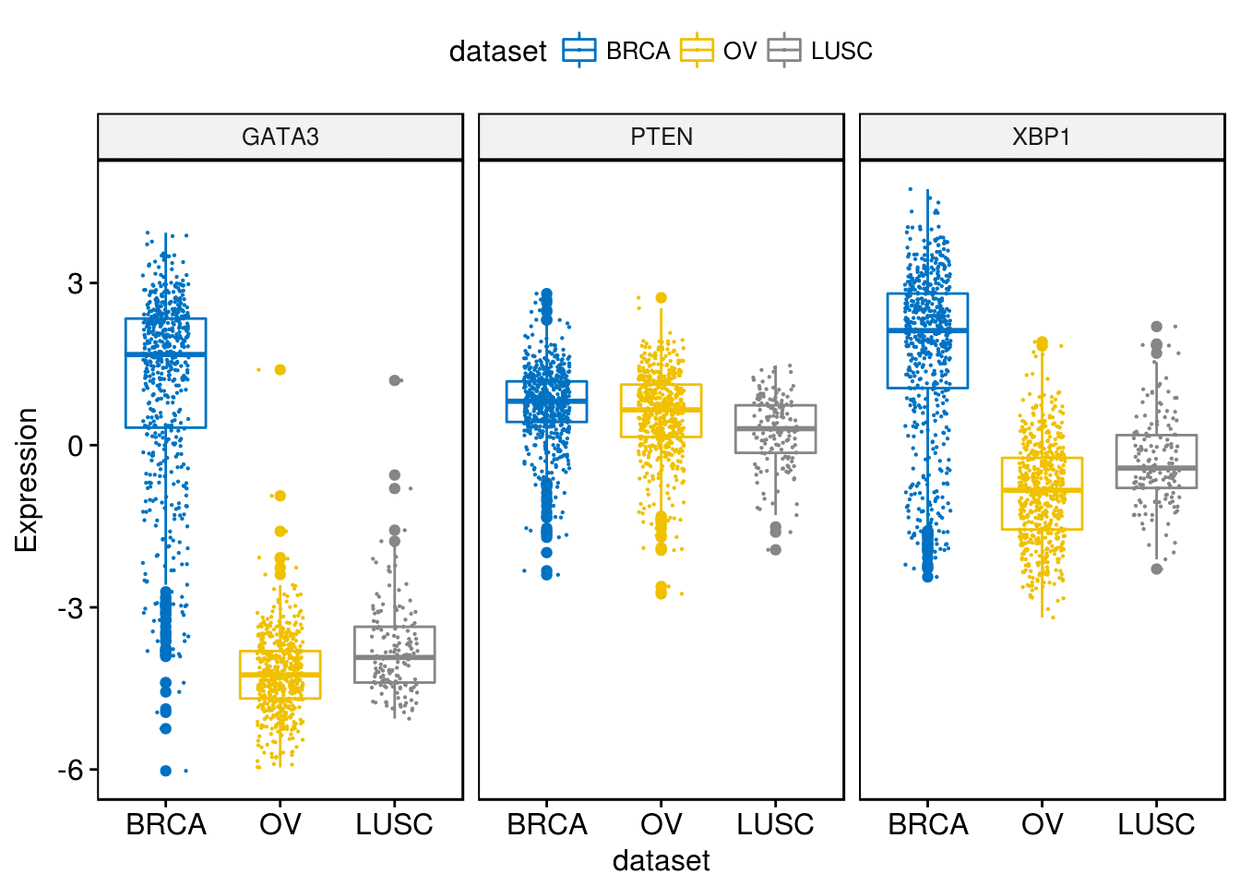
 通过add=jitter增加抖动点
通过add=jitter增加抖动点
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", ylab = "Expression", add = "jitter",
add.params = list(size=0.1, jitter=0.2))
 通过add=dotplot增加dotplot
通过add=dotplot增加dotplot
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"),
combine = TRUE, color = "dataset", palette = "jco", ylab = "Expression",
add = "dotplot", add.params = list(binwidth=0.1, dotsize=0.2))
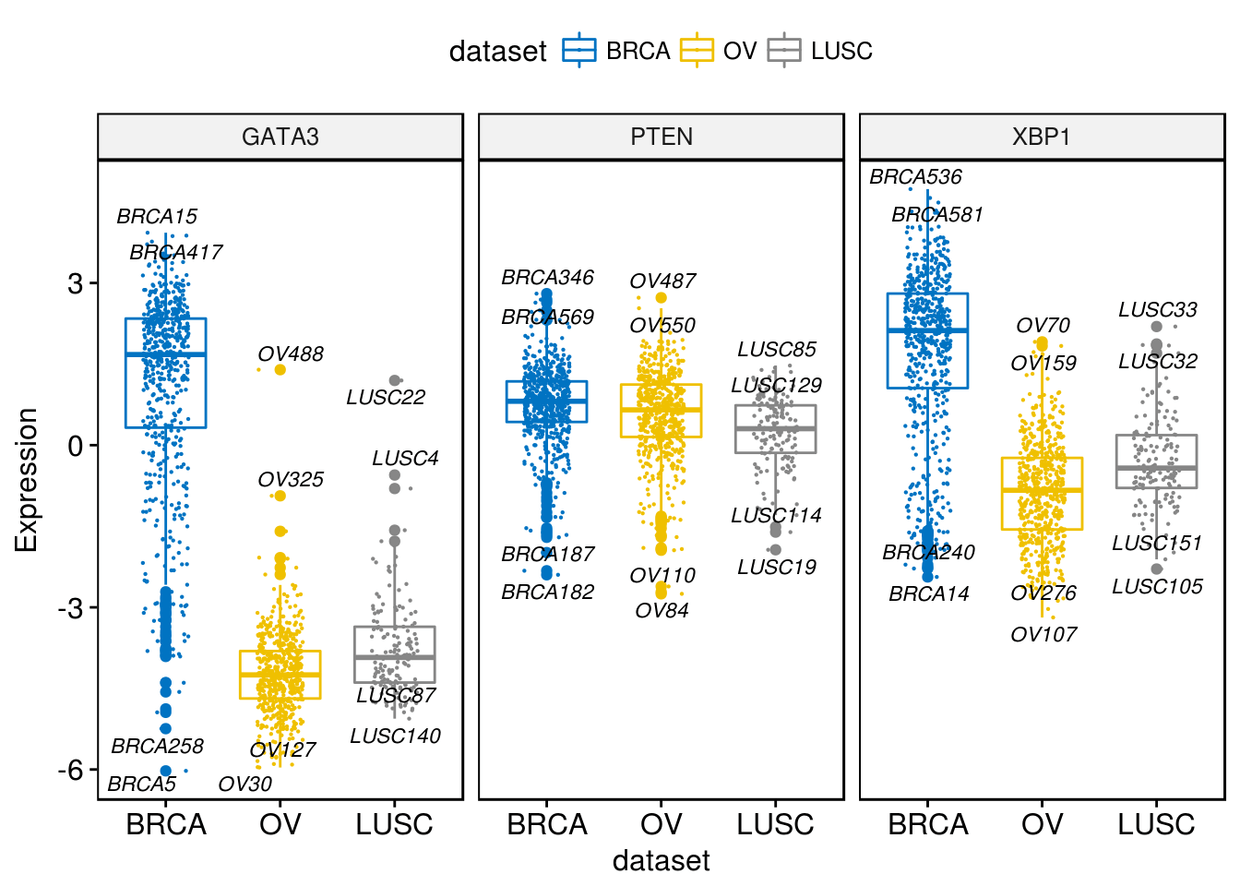
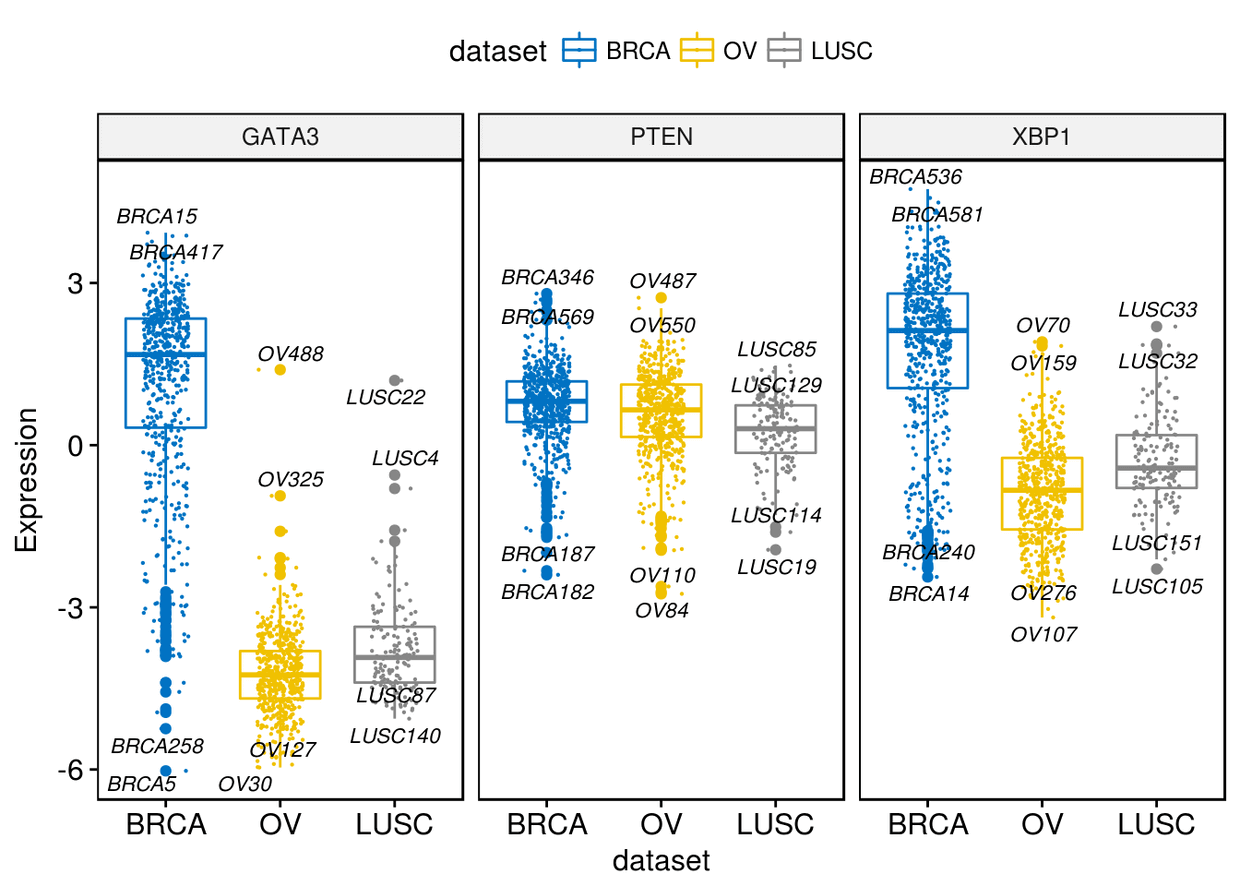
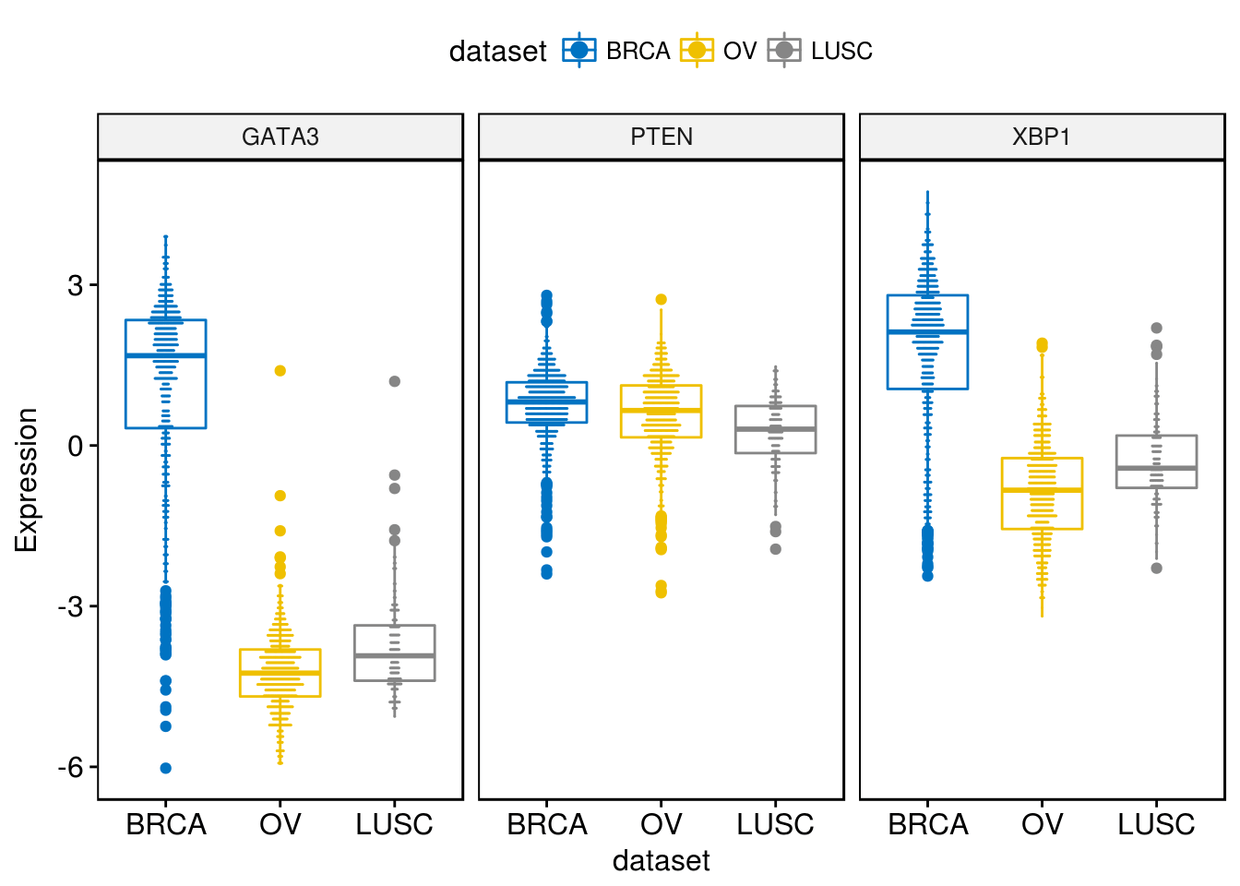 很多时候我们很像知道箱线图两端的数据,我们可以通过label来进行展示
很多时候我们很像知道箱线图两端的数据,我们可以通过label来进行展示
ggboxplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", ylab = "Expression", add = "jitter",
add.params = list(size=0.1, jitter=0.2), label = "bcr_patient_barcode",
label.select = list(top.up=2, top.down=2),
font.label = list(size=9, face="italic"), repel = TRUE)
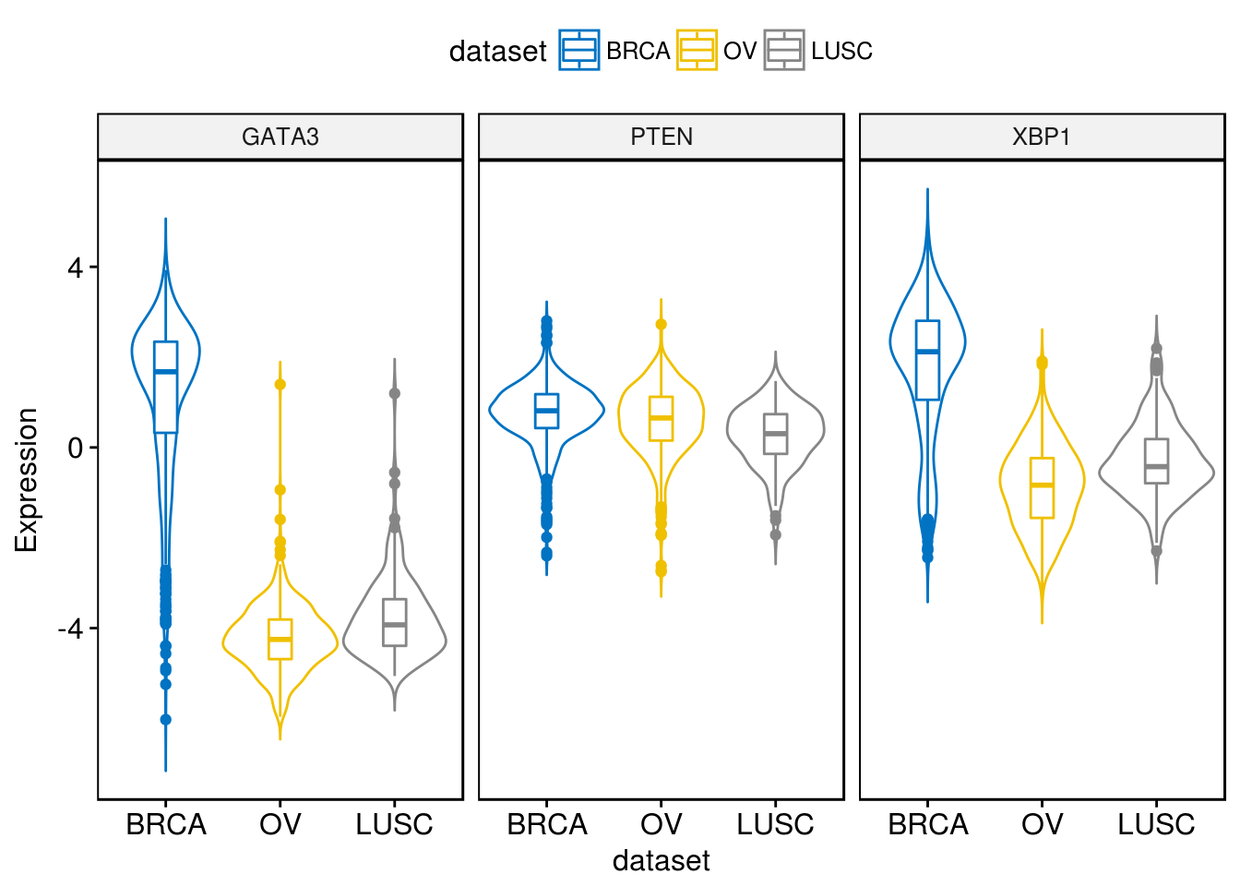
 2、小提琴图
2、小提琴图
ggviolin(expr,x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", ylab = "Expression", add = "boxplot")
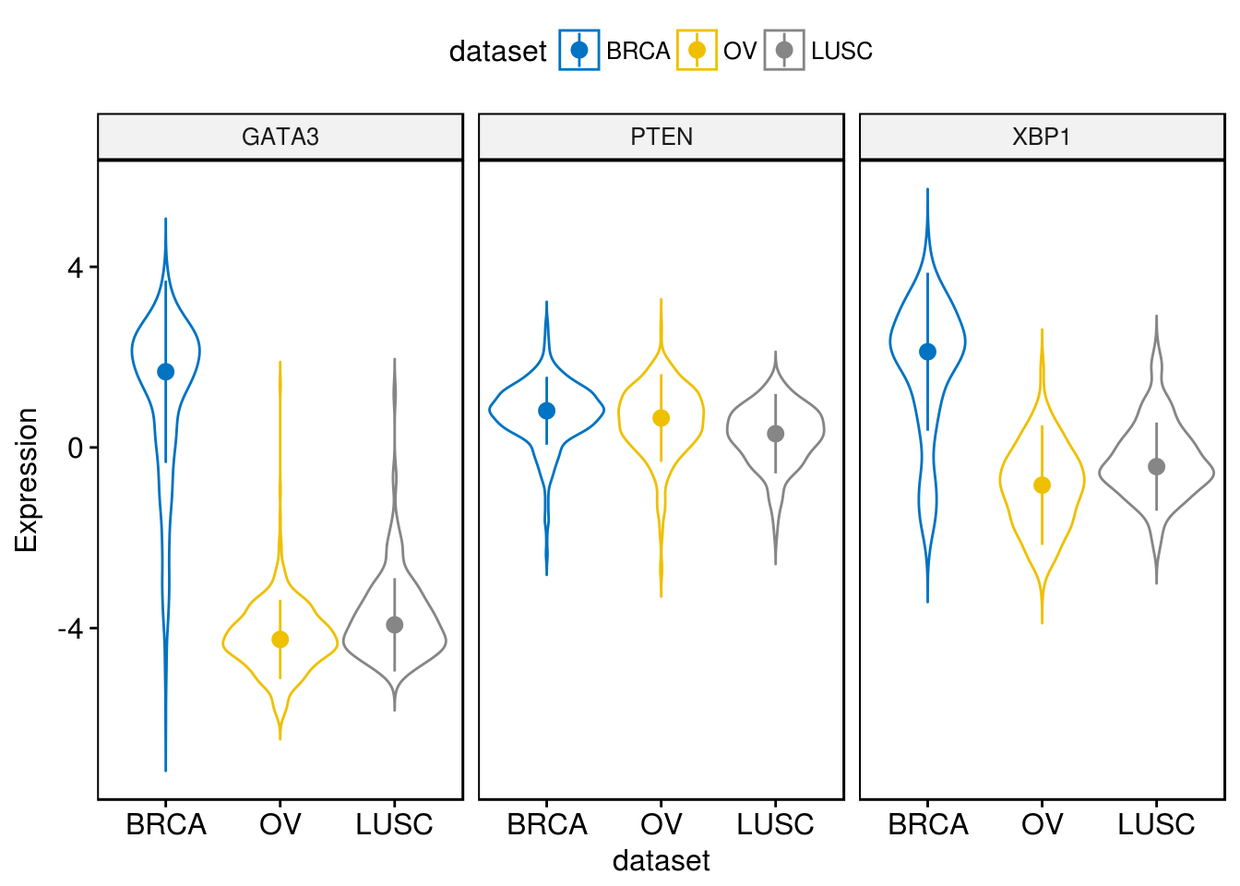
 通过修改add来更改添加小提琴图里的图形
通过修改add来更改添加小提琴图里的图形
ggviolin(expr,x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", ylab = "Expression", add = "median_iqr")
add有好多选项可以选择:“mean”, “mean_se”, “mean_sd”, “mean_ci”, “mean_range”, “median”, “median_iqr”, “median_mad”, “median_range”.有兴趣的可以自己试试。
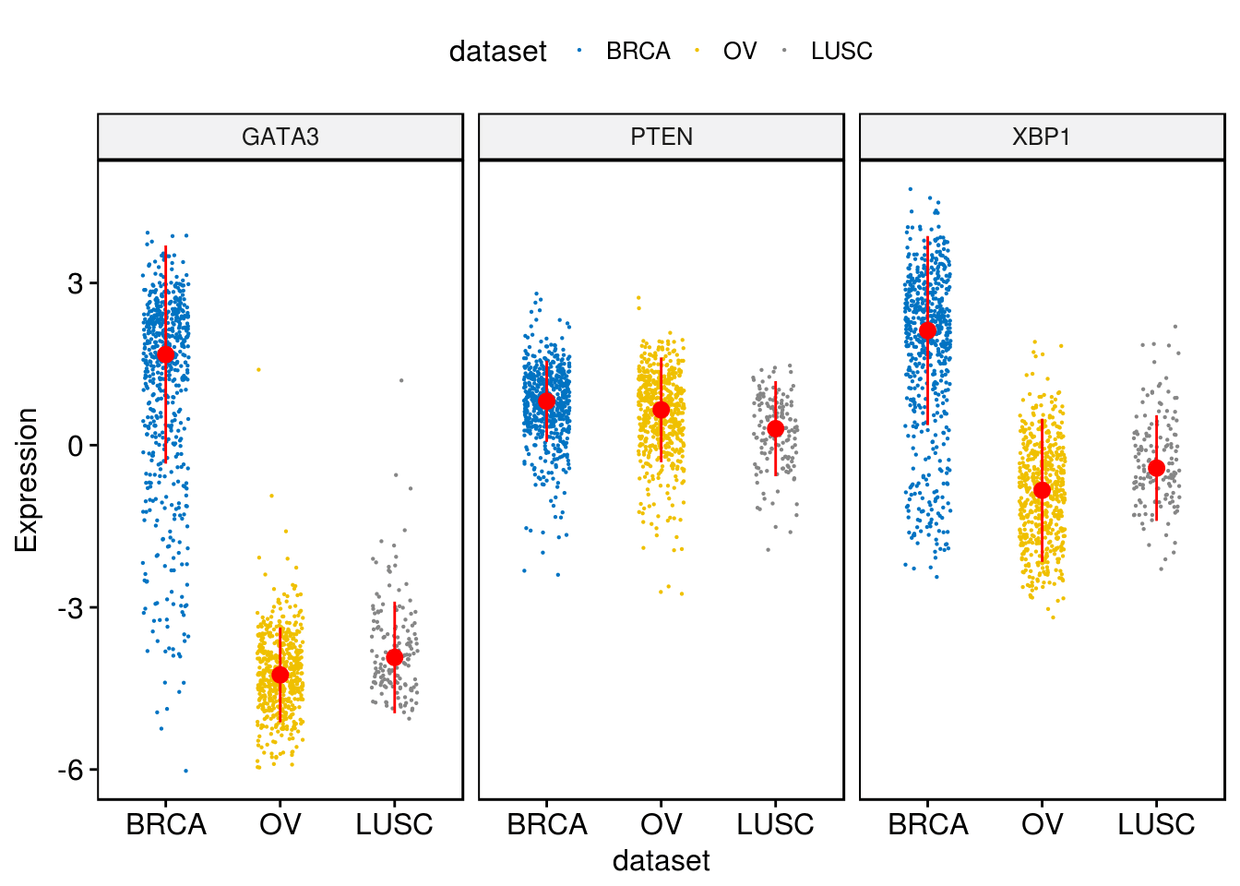
3、带状图
ggstripchart(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", size = 0.1, jitter=0.2, ylab = "Expression",
add = "median_iqr", add.params = list(color="red"))
 4、dotplot
4、dotplot
ggdotplot(expr, x="dataset", y=c("GATA3", "PTEN", "XBP1"), combine = TRUE,
color = "dataset", palette = "jco", fill = "white", binwidth = 0.1, ylab = "Expression",
add = "median_iqr", add.params = list(size=0.9))
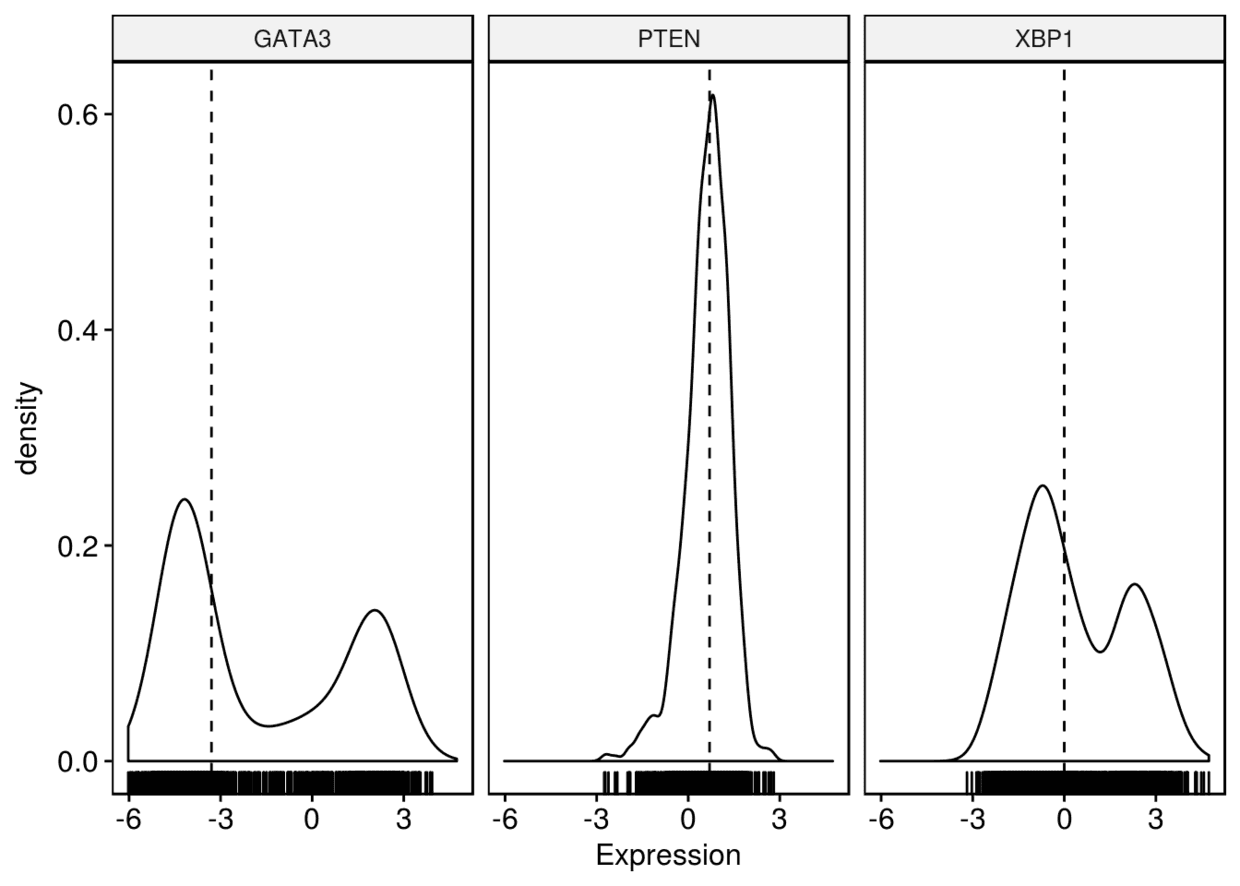
 5、密度图
5、密度图
ggdensity(expr, x=c("GATA3", "PTEN", "XBP1"), y="..density..",
combine = TRUE, xlab = "Expression", add = "median", rug = TRUE)
 将dataset映射给颜色
将dataset映射给颜色
ggdensity(expr, x=c("GATA3", "PTEN", "XBP1"), y="..density..", combine = TRUE,
xlab = "Expression", add = "median", rug = TRUE, color = "dataset",
fill = "dataset", palette = "jco")
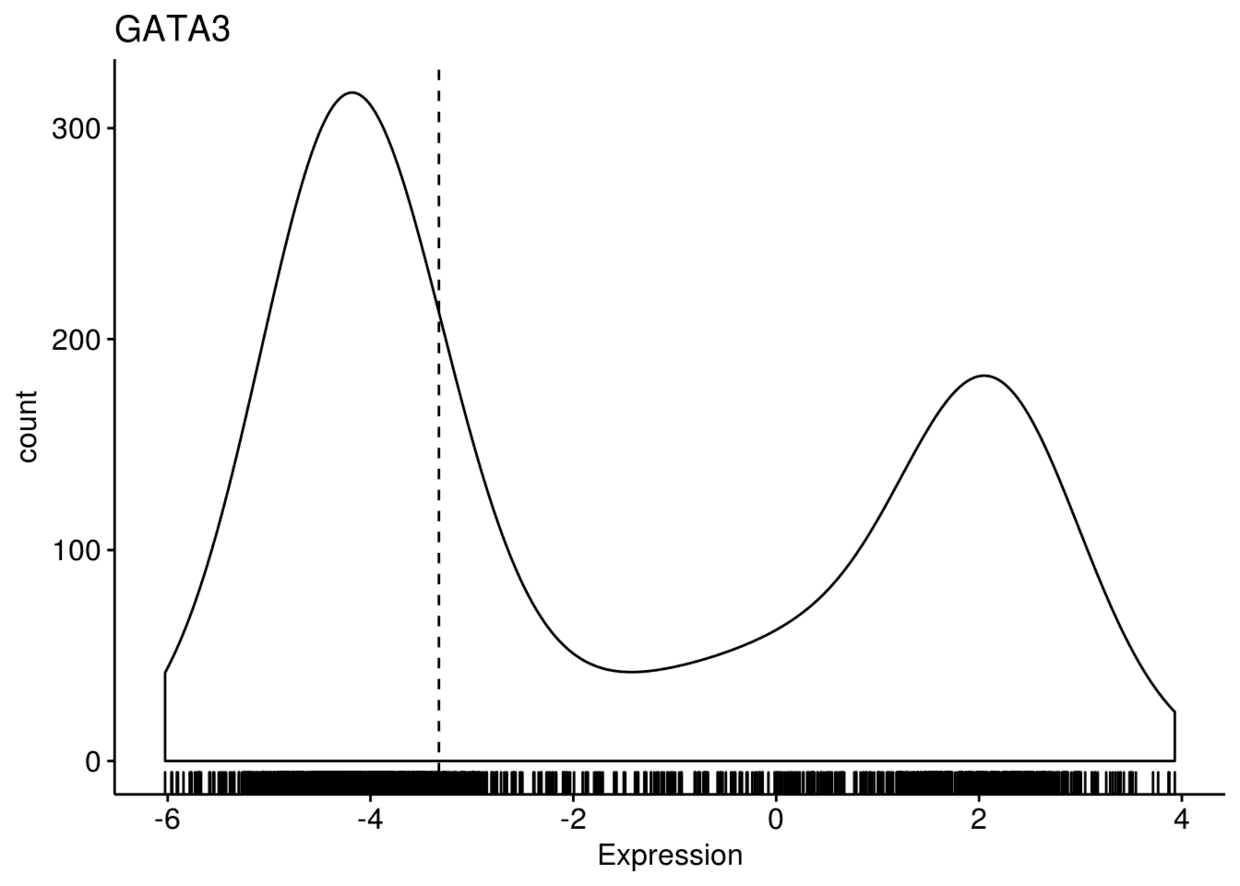
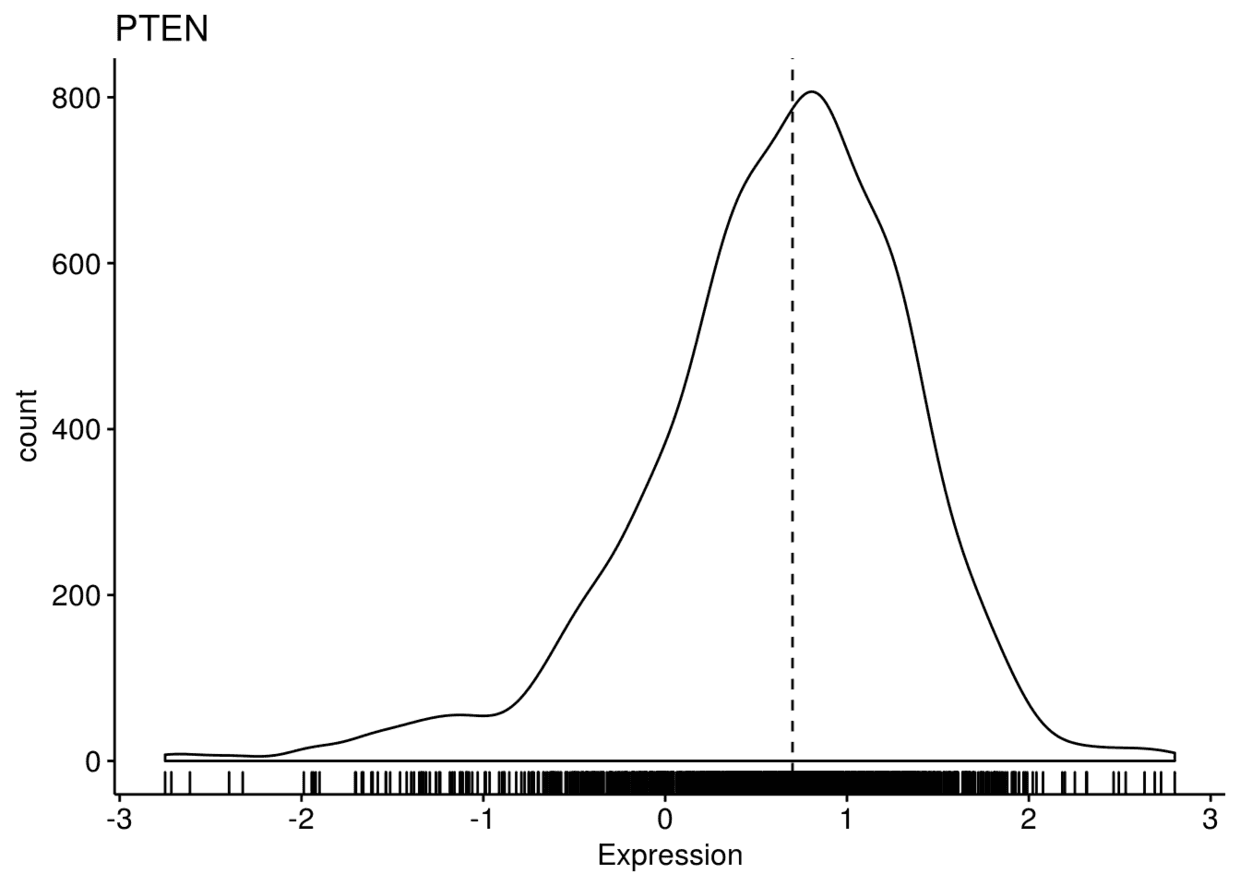
 将三幅图整合进一个panel中,并对y轴进行..count..,而不是..density..
将三幅图整合进一个panel中,并对y轴进行..count..,而不是..density..
ggdensity(expr, x=c("GATA3", "PTEN", "XBP1"), y="..count..", xlab = "Expression",
add = "median", rug = TRUE, palette = "jco")
## $GATA3
## ## $PTEN
## ## $XBP1
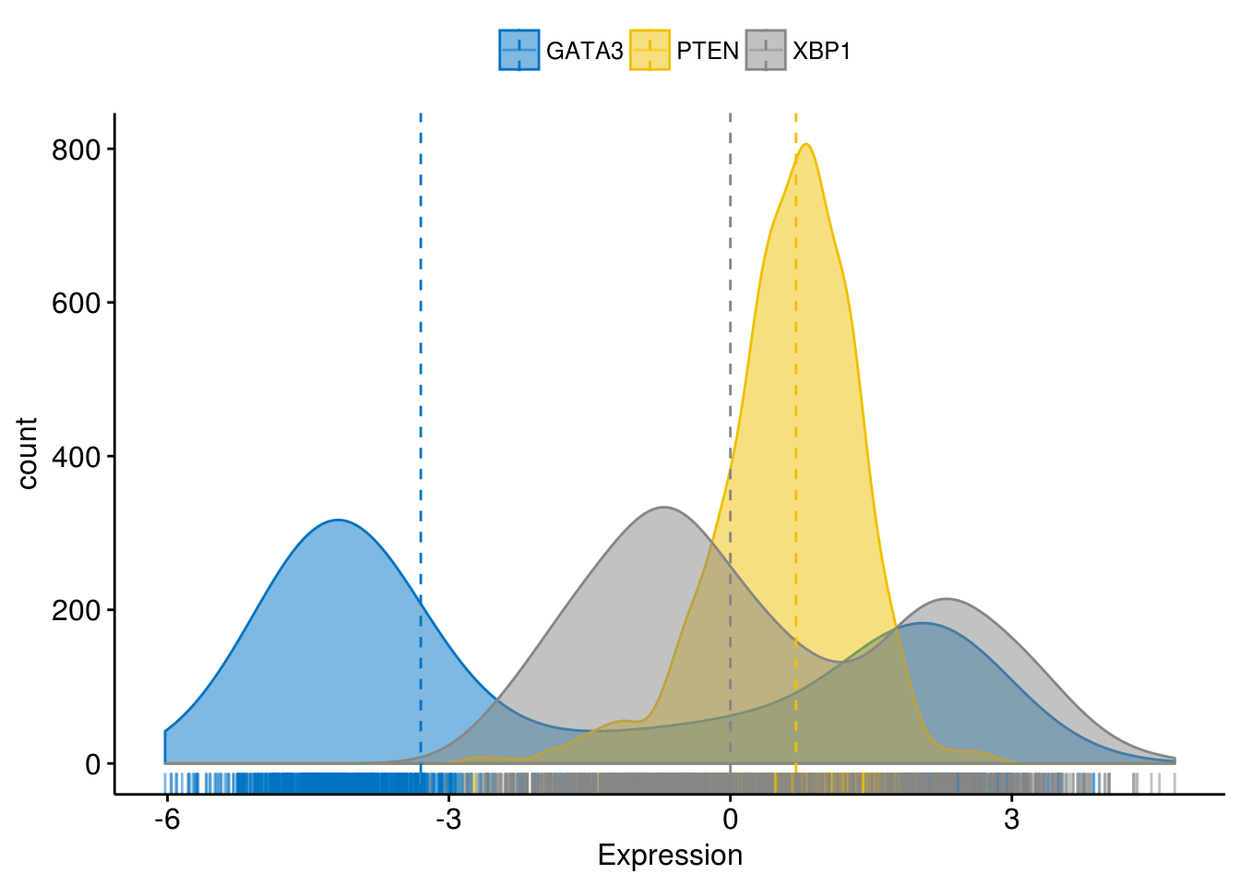
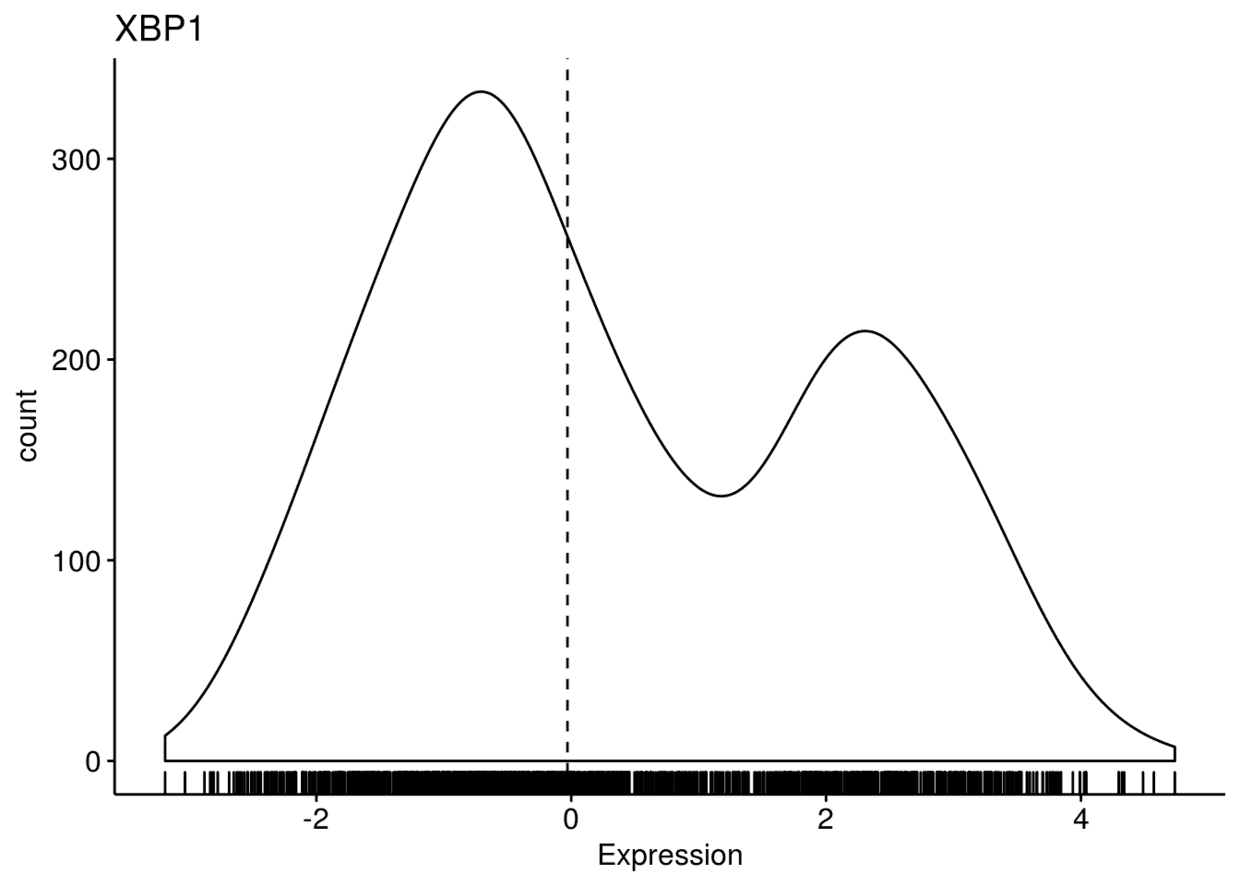 颜色映射,将x轴变量映射给颜色
颜色映射,将x轴变量映射给颜色
ggdensity(expr, x=c("GATA3", "PTEN", "XBP1"), y="..count..", color = ".x.",
fill = ".x.", merge = TRUE, xlab = "Expression", add = "median", rug = TRUE, palette = "jco")
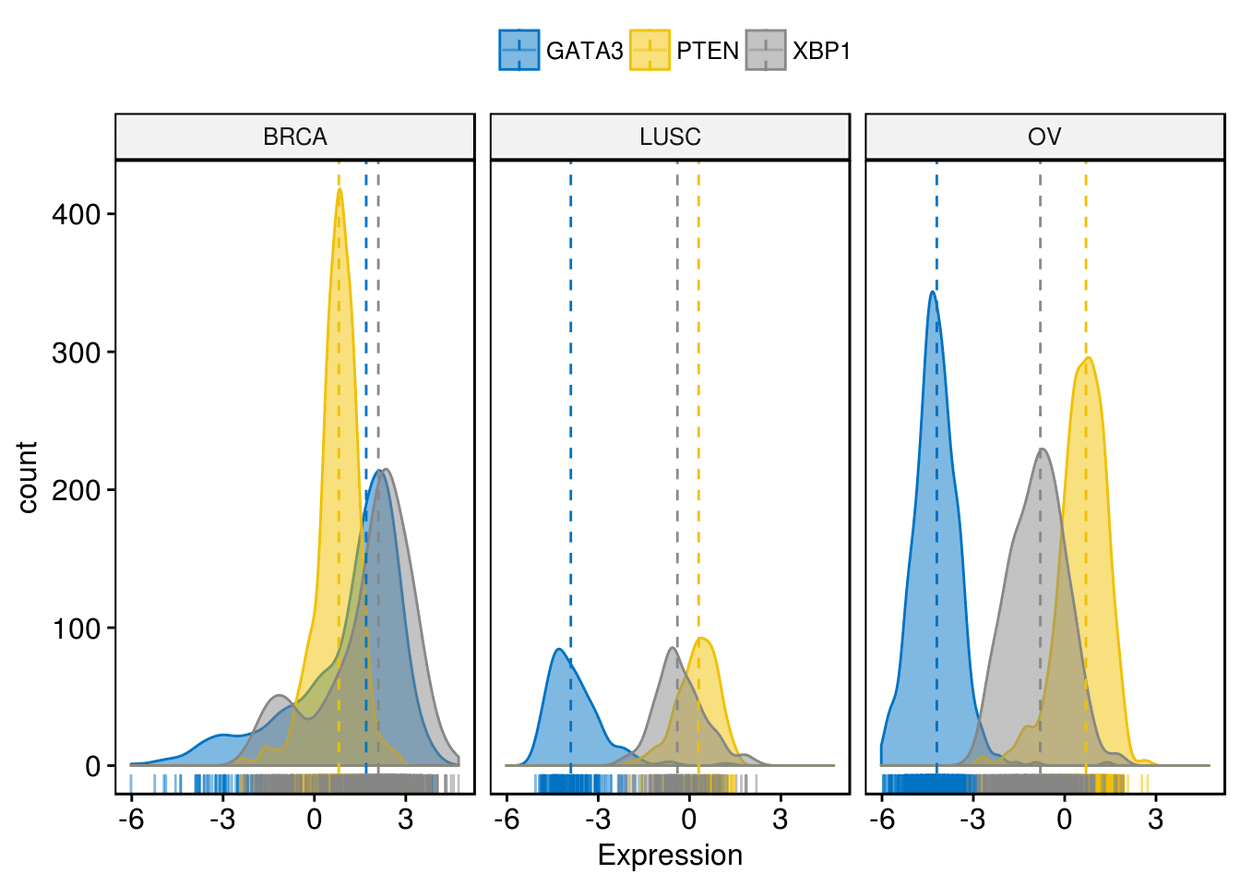
 按dataset进行分面
按dataset进行分面
ggdensity(expr, x=c("GATA3", "PTEN", "XBP1"), y="..count..", color = ".x.",
fill = ".x.", merge = TRUE, xlab = "Expression", add = "median",
rug = TRUE, palette = "jco", facet.by = "dataset")
 6、直方图
6、直方图
gghistogram(expr, x=c("GATA3", "PTEN", "XBP1"), y="..density..",
xlab = "Expression", add = "median", rug = TRUE)
## $GATA3
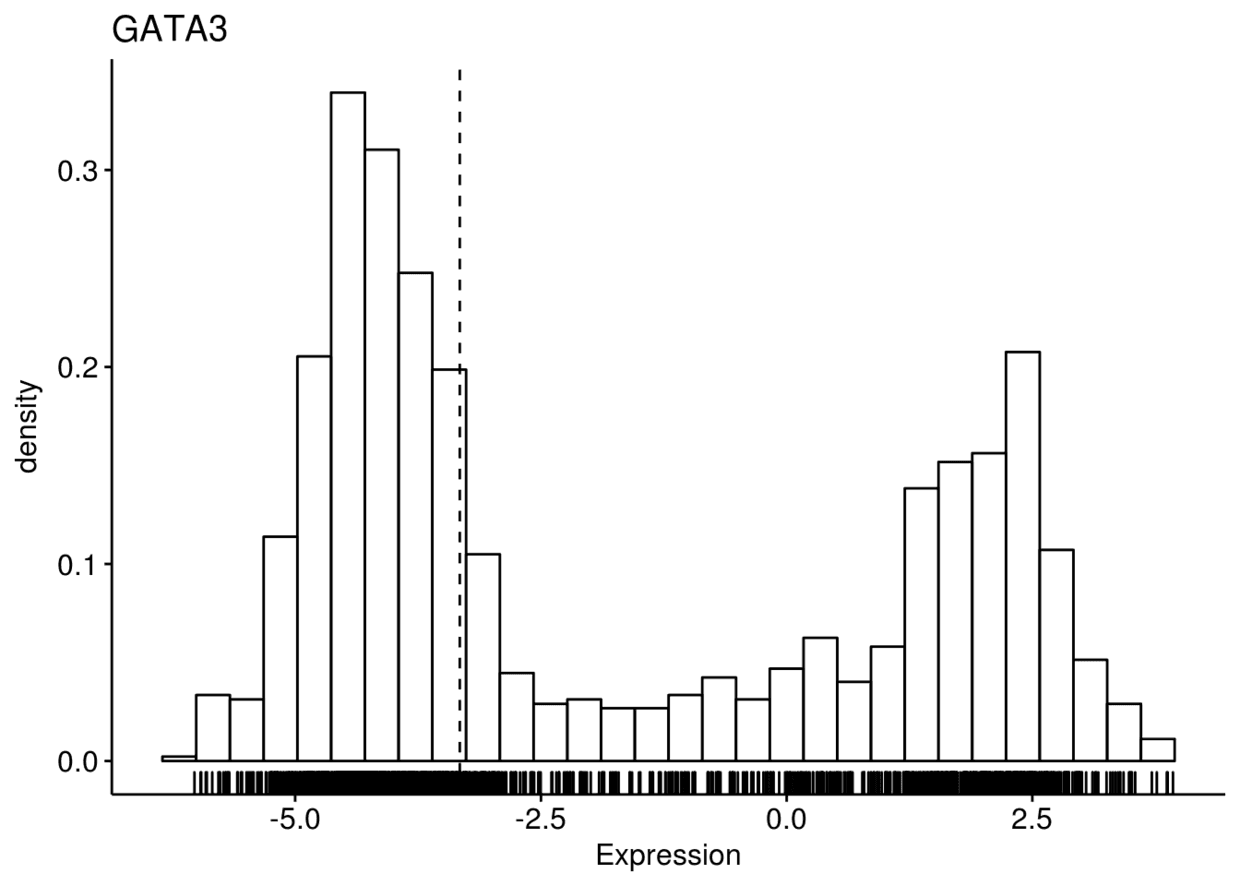
## ## $PTEN
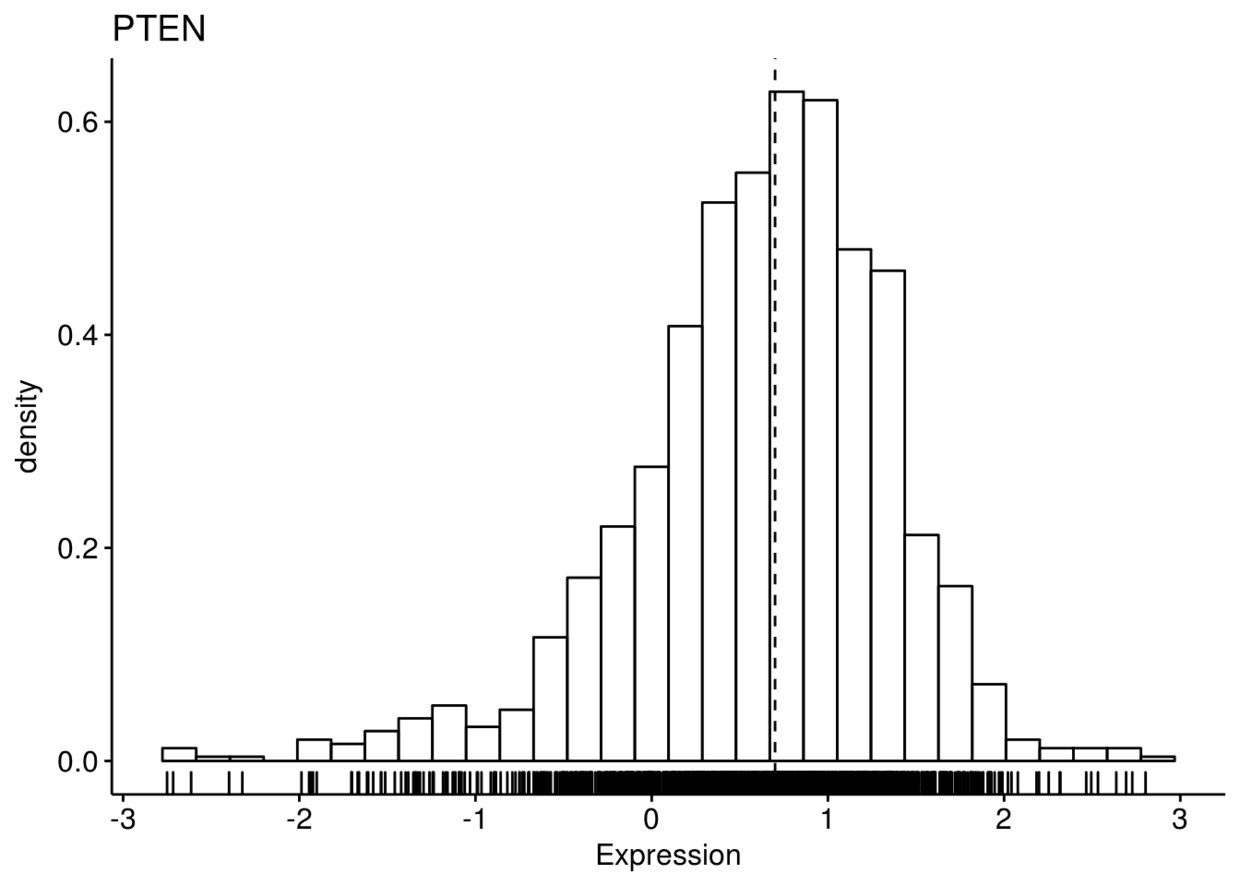
## ## $XBP1
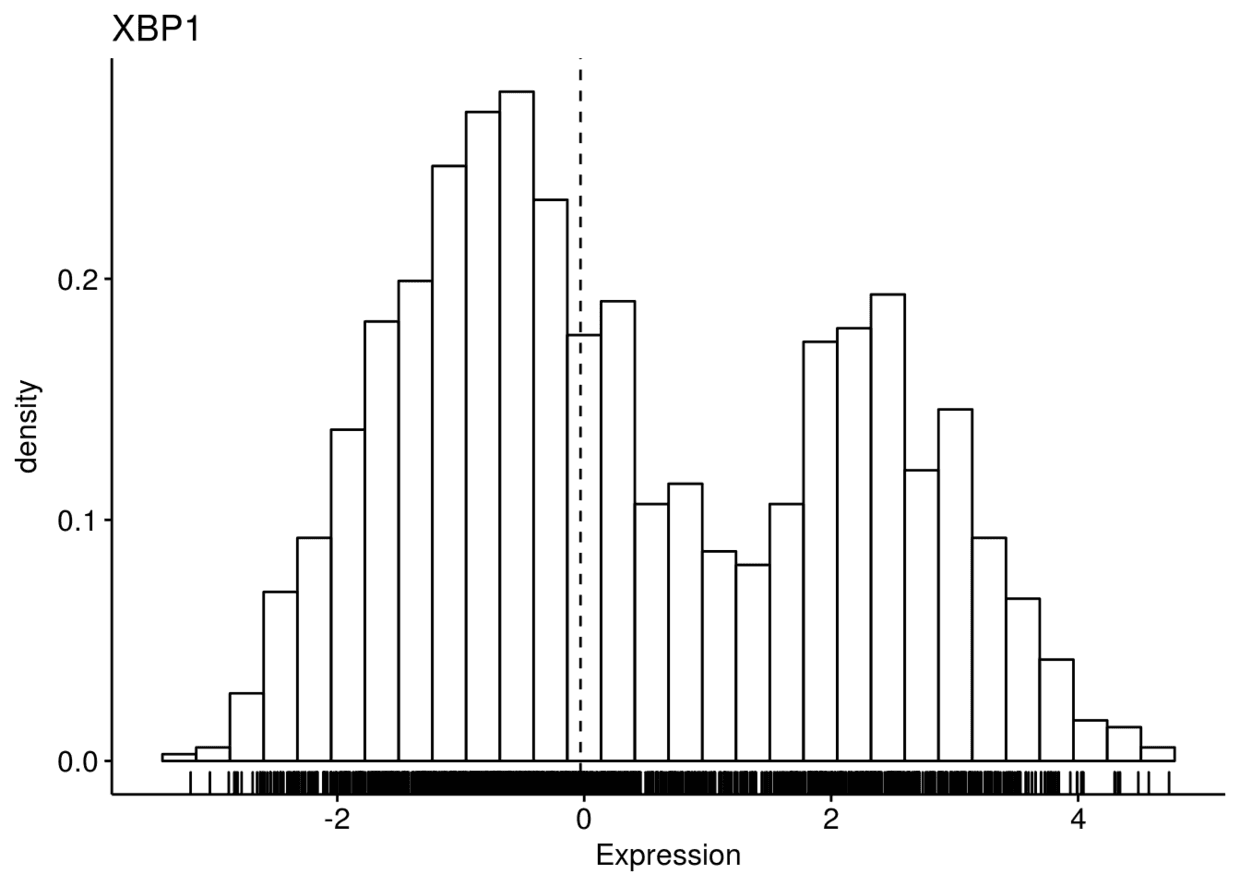 将dataset映射给颜色
将dataset映射给颜色
gghistogram(expr, x=c("GATA3", "PTEN", "XBP1"), y="..density..", xlab = "Expression",
add = "median", rug = TRUE, color = "dataset", fill = "dataset", palette = "jco")
## $GATA3
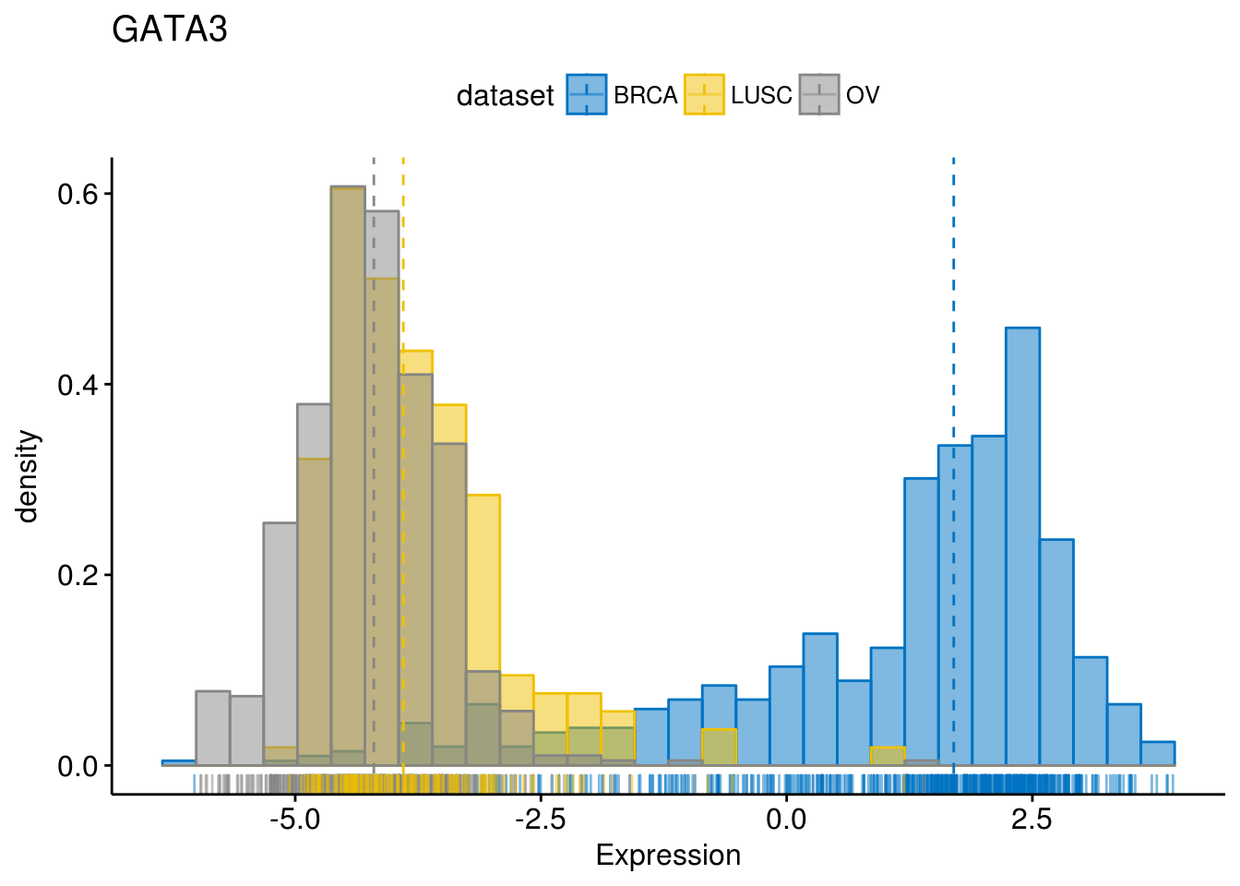
## ## $PTEN
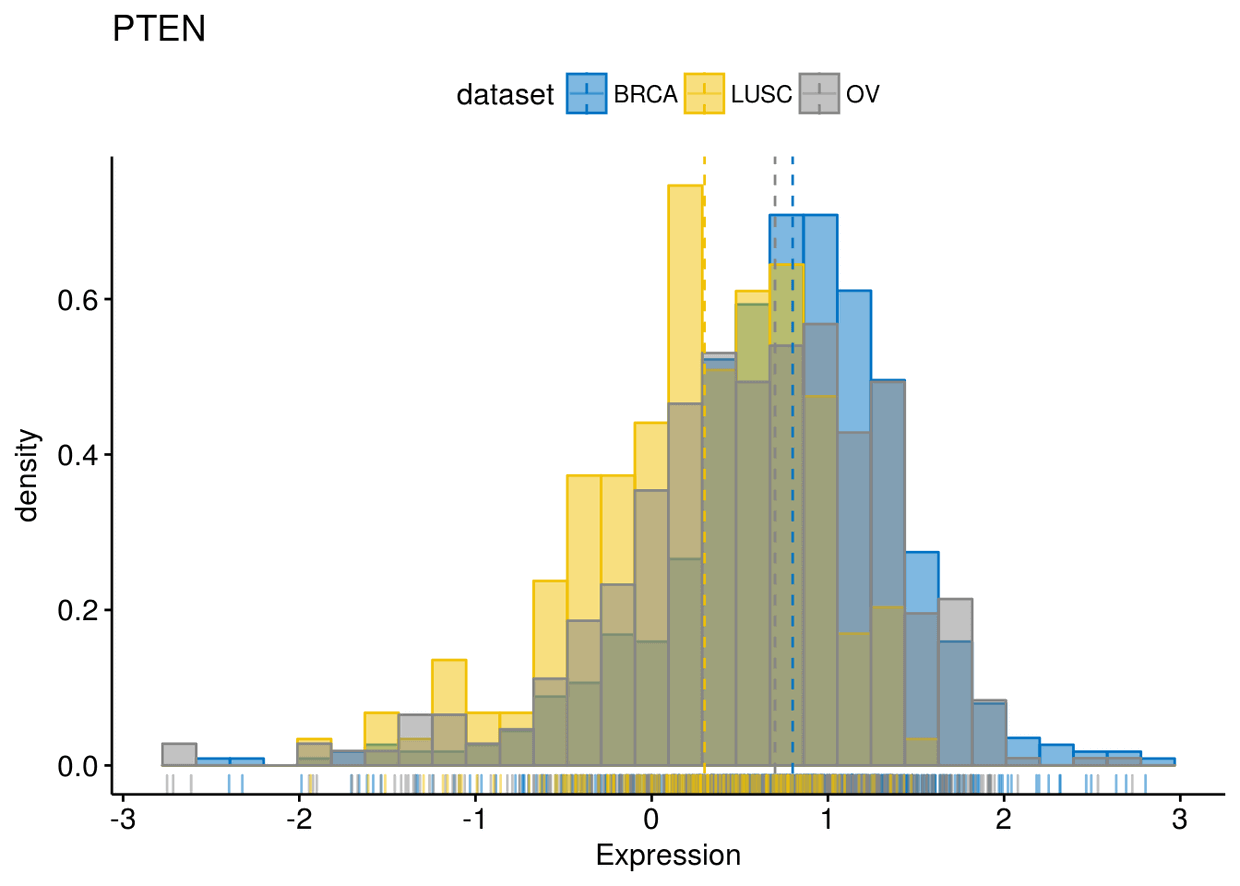
## ## $XBP1
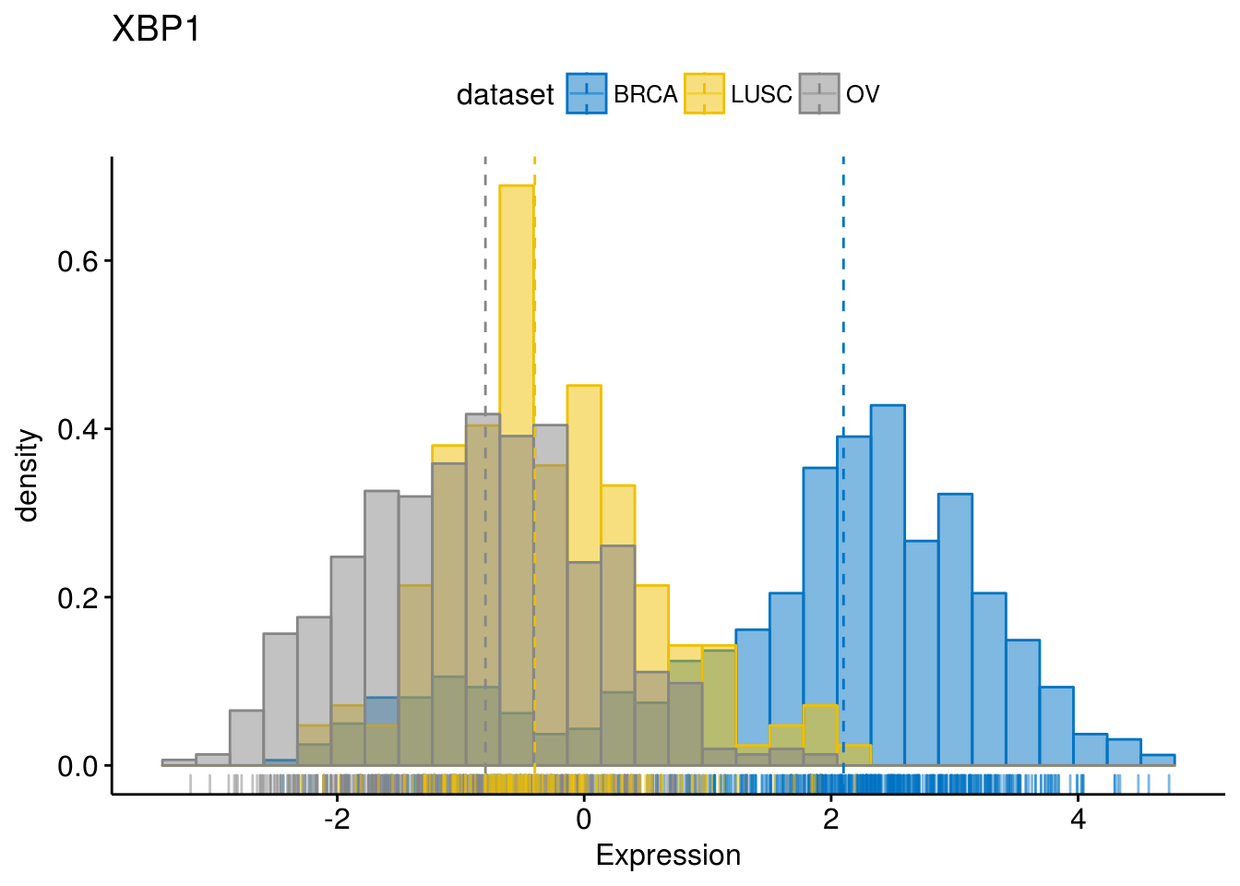 后面还有一些将几幅图整合在一个panel以及分面等大同小异就不讲了。
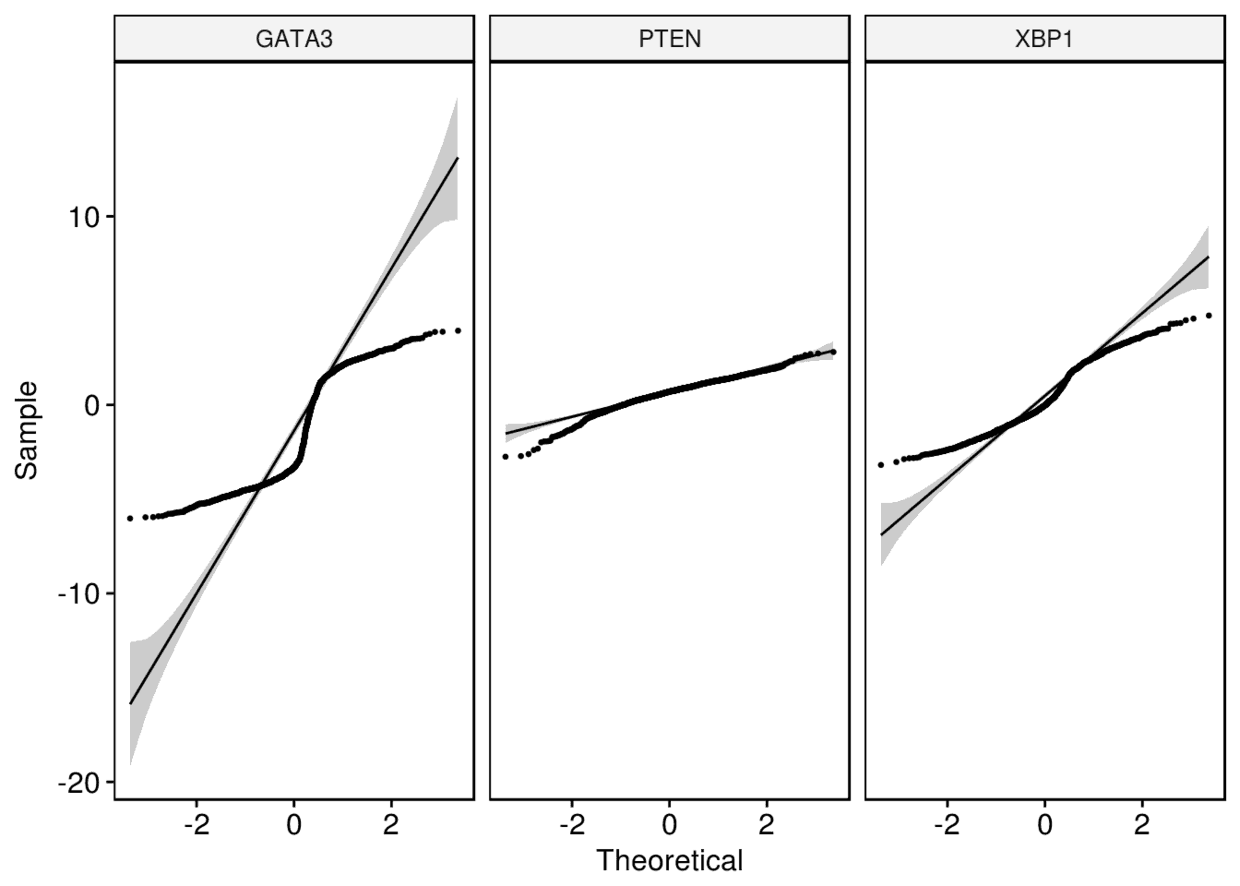
7、Q-Q图
后面还有一些将几幅图整合在一个panel以及分面等大同小异就不讲了。
7、Q-Q图
ggqqplot(expr, x=c("GATA3", "PTEN", "XBP1"), combine = TRUE, size = 0.5)
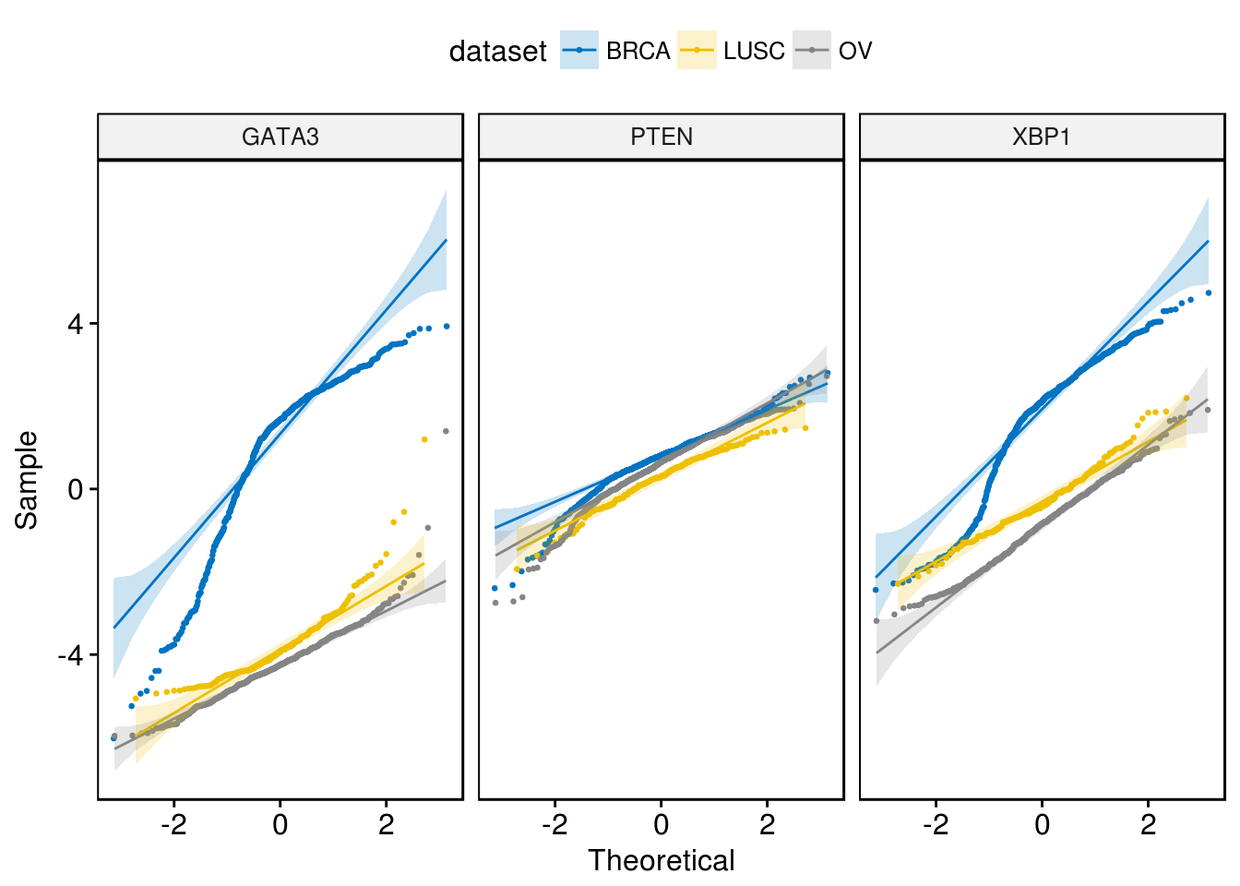
 颜色映射
颜色映射
ggqqplot(expr, x=c("GATA3", "PTEN", "XBP1"), combine = TRUE, size = 0.5,
color = "dataset", palette = "jco")
#sessionInfo
sessionInfo()
## R version 3.4.0 (2017-04-21)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 16.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/libblas/libblas.so.3.6.0
## LAPACK: /usr/lib/lapack/liblapack.so.3.6.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=zh_CN.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=zh_CN.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=zh_CN.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] bindrcpp_0.2 RTCGA.mRNA_1.4.0 RTCGA_1.6.0 ggpubr_0.1.3
## [5] magrittr_1.5 ggplot2_2.2.1
##
## loaded via a namespace (and not attached):
## [1] zoo_1.8-0 reshape2_1.4.2 purrr_0.2.2.2
## [4] splines_3.4.0 ggthemes_3.4.0 lattice_0.20-35
## [7] colorspace_1.3-2 htmltools_0.3.6 viridisLite_0.2.0
## [10] yaml_2.1.14 survival_2.41-3 XML_3.98-1.9
## [13] survMisc_0.5.4 rlang_0.1.1 foreign_0.8-68
## [16] glue_1.1.0 bindr_0.1 plyr_1.8.4
## [19] stringr_1.2.0 ggsignif_0.2.0 munsell_0.4.3
## [22] gtable_0.2.0 ggsci_2.7 rvest_0.3.2
## [25] psych_1.7.5 evaluate_0.10 labeling_0.3
## [28] knitr_1.16 parallel_3.4.0 broom_0.4.2
## [31] Rcpp_0.12.11 xtable_1.8-2 scales_0.4.1
## [34] backports_1.1.0 cmprsk_2.2-7 km.ci_0.5-2
## [37] gridExtra_2.2.1 mnormt_1.5-5 digest_0.6.12
## [40] stringi_1.1.5 ggrepel_0.6.5 dplyr_0.7.0
## [43] KMsurv_0.1-5 grid_3.4.0 rprojroot_1.2
## [46] tools_3.4.0 lazyeval_0.2.0 tibble_1.3.3
## [49] tidyr_0.6.3 Matrix_1.2-10 data.table_1.10.4
## [52] xml2_1.1.1 survminer_0.4.0 assertthat_0.2.0
## [55] rmarkdown_1.6 httr_1.2.1 viridis_0.4.0
## [58] R6_2.2.2 nlme_3.1-131 compiler_3.4.0