R语言学习笔记之聚类分析
使用k-means聚类所需的包:
- factoextra
- cluster
加载包
library(factoextra)
library(cluster)
数据准备
使用内置的R数据集USArrests
#load the dataset
data("USArrests")
#remove any missing value (i.e, NA values for not available)
#That might be present in the data
USArrests <- na.omit(USArrests)#view the first 6 rows of the data
head(USArrests, n=6)
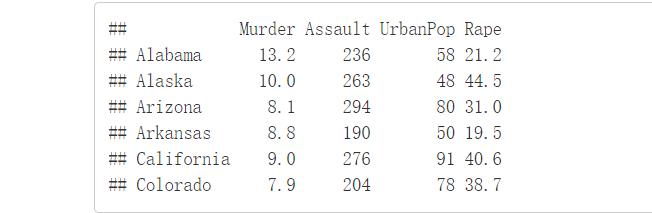
在此数据集中,列是变量,行是观测值 在聚类之前我们可以先进行一些必要的数据检查即数据描述性统计,如平均值、标准差等
desc_stats <- data.frame( Min=apply(USArrests, 2, min),#minimum
Med=apply(USArrests, 2, median),#median
Mean=apply(USArrests, 2, mean),#mean
SD=apply(USArrests, 2, sd),#Standard deviation
Max=apply(USArrests, 2, max)#maximum
)
desc_stats <- round(desc_stats, 1)#保留小数点后一位head(desc_stats)
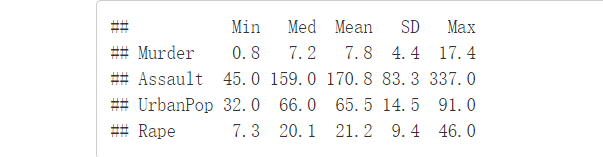
变量有很大的方差及均值时需进行标准化
df <- scale(USArrests)
数据集群性评估
使用get_clust_tendency()计算Hopkins统计量
res <- get_clust_tendency(df, 40, graph = TRUE)
res$hopkins_stat
## [1] 0.3440875
#Visualize the dissimilarity matrix
res$plot

Hopkins统计量的值<0.5,表明数据是高度可聚合的。另外,从图中也可以看出数据可聚合。
估计聚合簇数
由于k均值聚类需要指定要生成的聚类数量,因此我们将使用函数clusGap()来计算用于估计最优聚类数。函数fviz_gap_stat()用于可视化。
set.seed(123)
## Compute the gap statistic
gap_stat <- clusGap(df, FUN = kmeans, nstart = 25, K.max = 10, B = 500)
# Plot the result
fviz_gap_stat(gap_stat)

图中显示最佳为聚成四类(k=4)
进行聚类
set.seed(123)
km.res <- kmeans(df, 4, nstart = 25)
head(km.res$cluster, 20)

# Visualize clusters using factoextra
fviz_cluster(km.res, USArrests)

检查cluster silhouette图
Recall that the silhouette measures (SiSi) how similar an object ii is to the the other objects in its own cluster versus those in the neighbor cluster. SiSi values range from 1 to - 1:
- A value of SiSi close to 1 indicates that the object is well clustered. In the other words, the object ii is similar to the other objects in its group.
- A value of SiSi close to -1 indicates that the object is poorly clustered, and that assignment to some other cluster would probably improve the overall results.
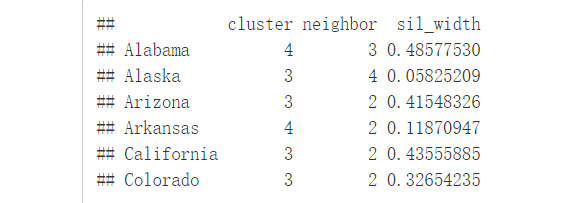
sil <- silhouette(km.res$cluster, dist(df))
rownames(sil) <- rownames(USArrests)
head(sil[, 1:3])
#Visualize
fviz_silhouette(sil)
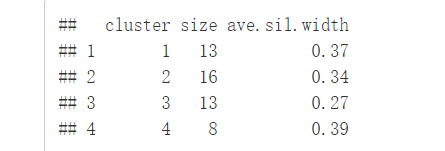
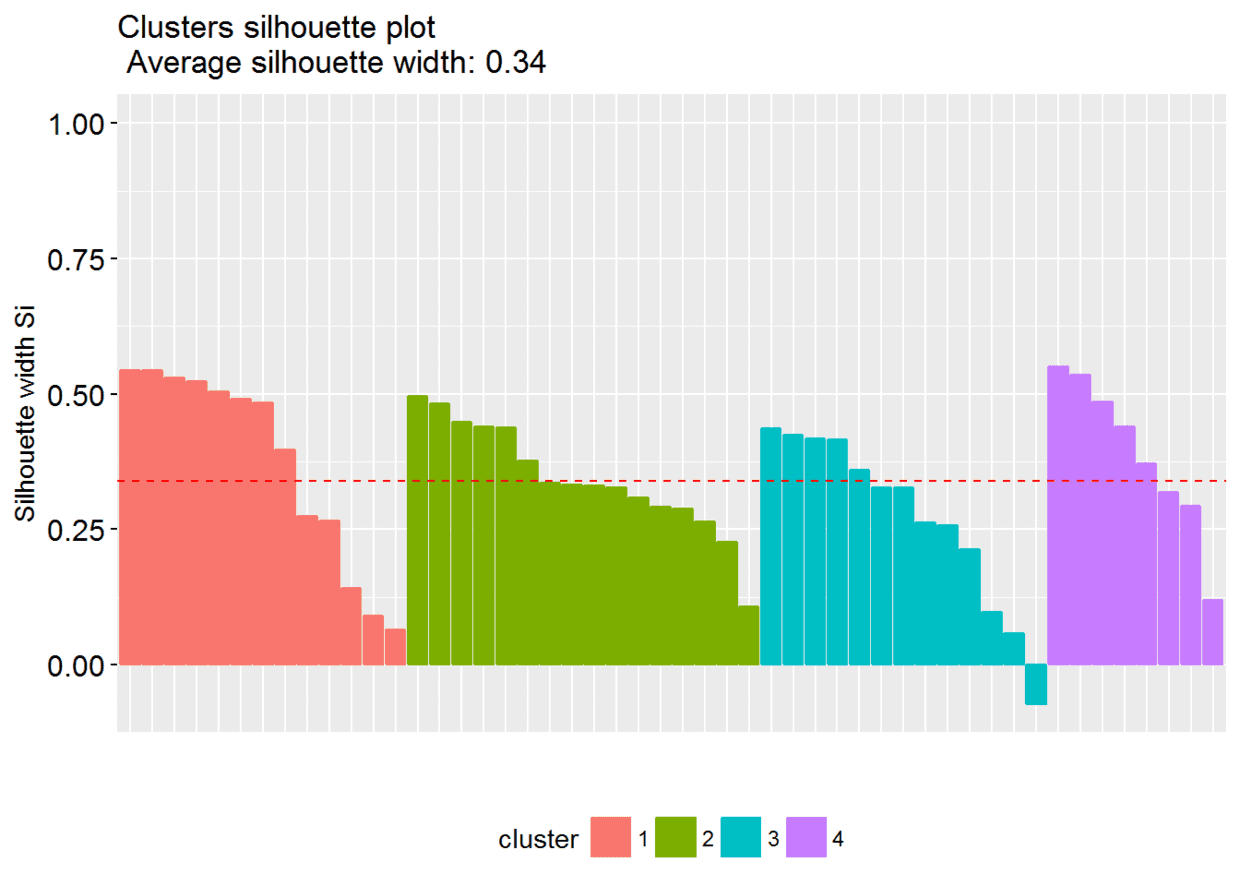 图中可以看出有负值,可以通过函数
图中可以看出有负值,可以通过函数silhouette()确定是哪个观测值
neg_sil_index <- which(sil[, "sil_width"] < 0)
sil[neg_sil_index, , drop = FALSE]
## cluster neighbor sil_width
## Missouri 3 2 -0.07318144
eclust():增强的聚类分析
与其他聚类分析包相比,
eclust()有以下优点:
- 简化了聚类分析的工作流程
- 可以用于计算层次聚类和分区聚类
- eclust()自动计算最佳聚类簇数。
- 自动提供Silhouette plot
- 可以结合ggplot2绘制优美的图形
使用eclust()的K均值聚类
# Compute k-means
res.km <- eclust(df, "kmeans")
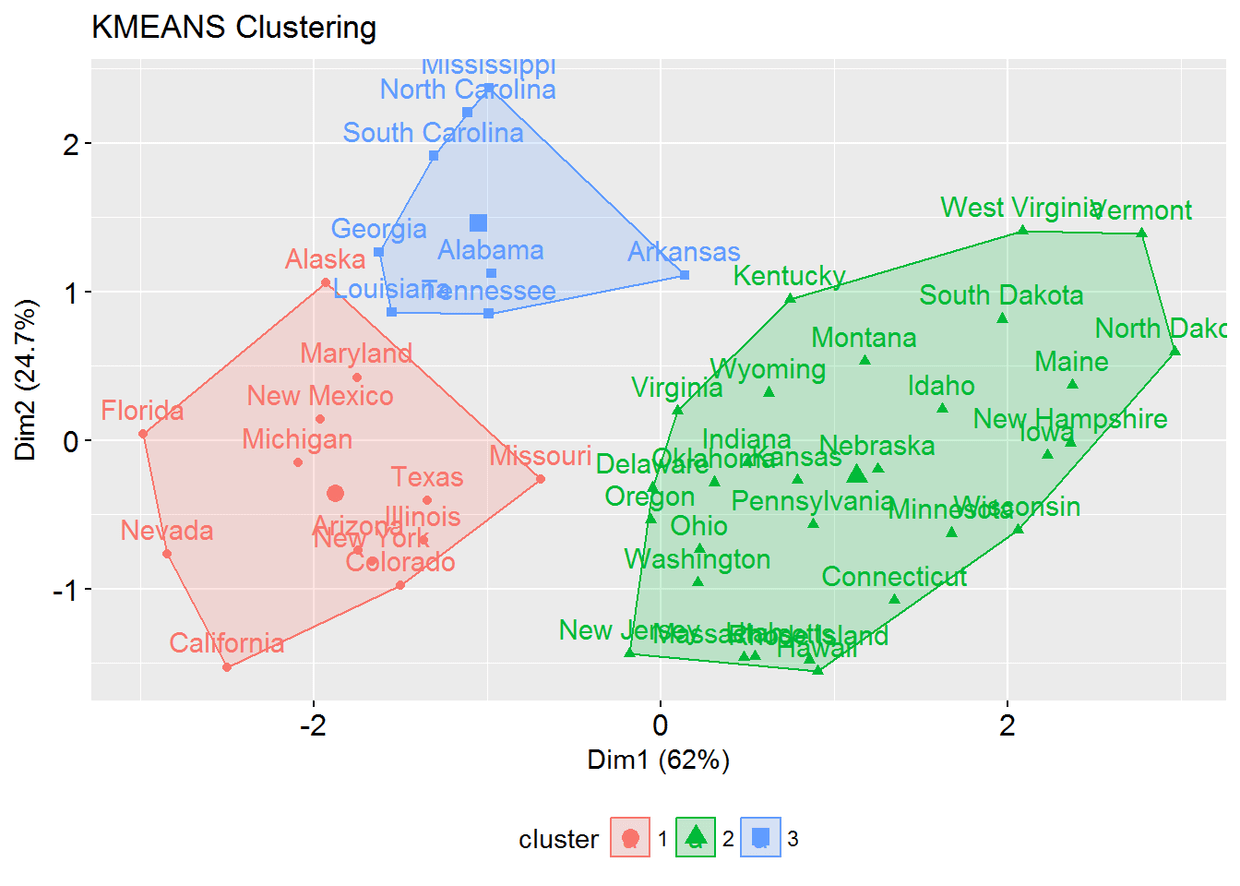
# Gap statistic plot
fviz_gap_stat(res.km$gap_stat)

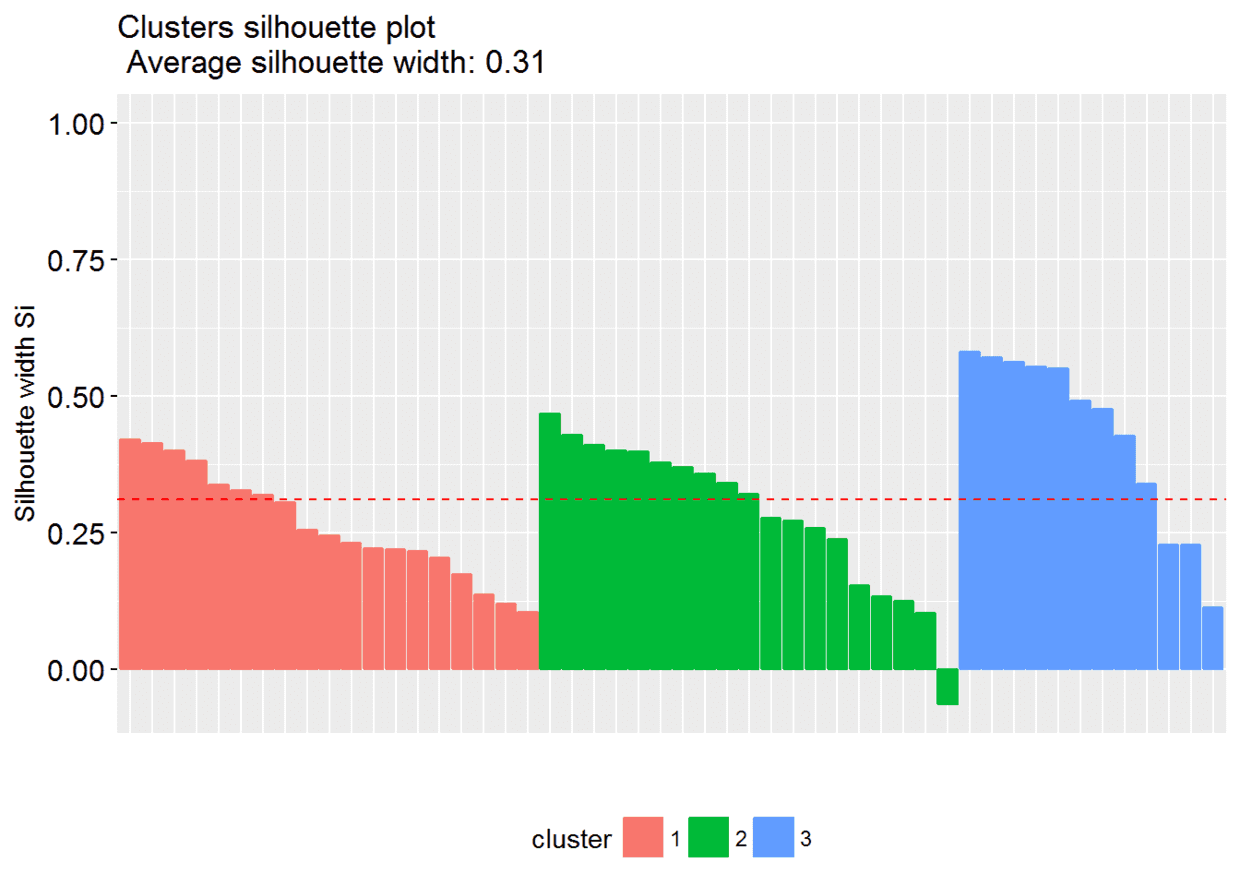
# Silhouette plotfviz_silhouette(res.km)
## cluster size ave.sil.width
## 1 1 13 0.31
## 2 2 29 0.38
## 3 3 8 0.39

使用**eclust()**的层次聚类
# Enhanced hierarchical clustering
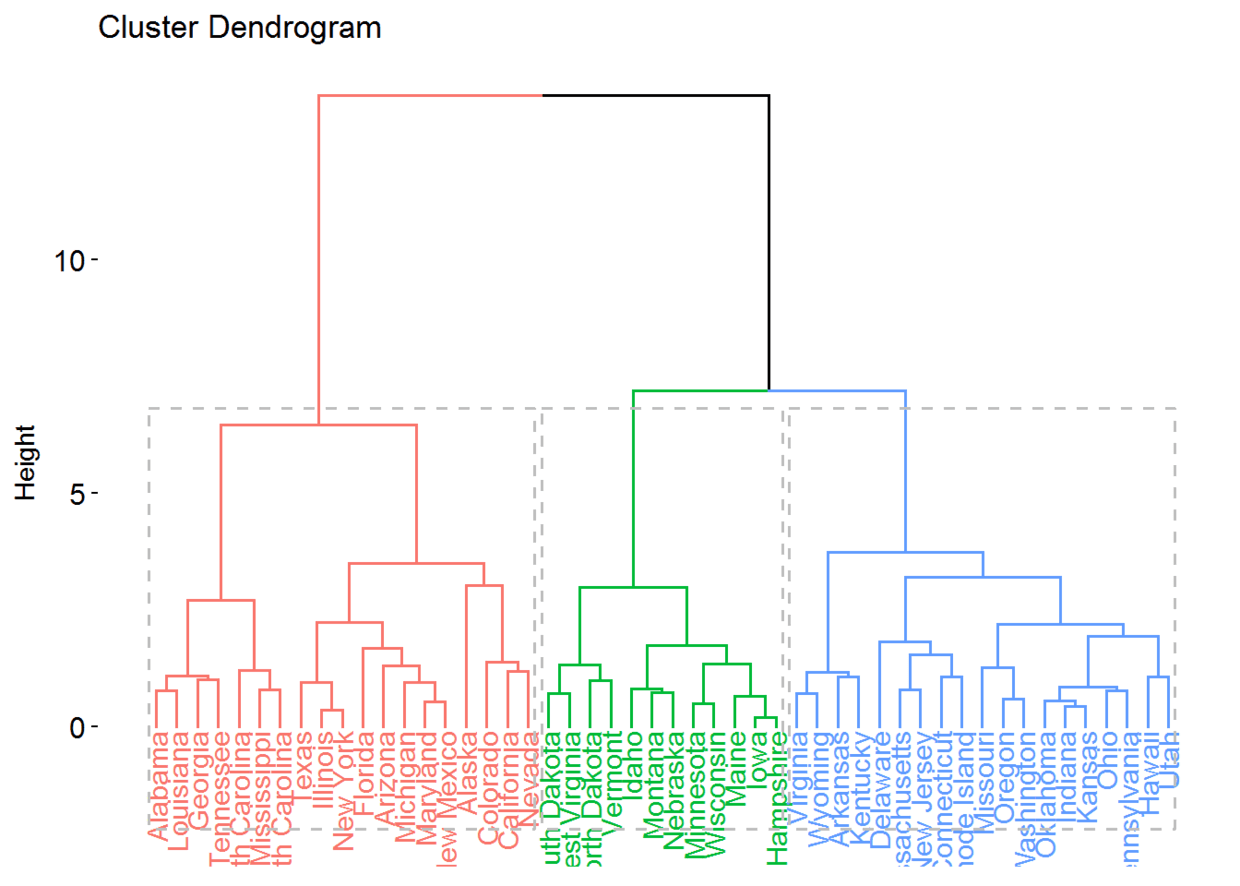
res.hc <- eclust(df, "hclust") # compute hclust
fviz_dend(res.hc, rect = TRUE) # dendrogam

#下面的R代码生成Silhouette plot和分层聚类散点图。
fviz_silhouette(res.hc) # silhouette plot
## cluster size ave.sil.width
## 1 1 19 0.26
## 2 2 19 0.28
## 3 3 12 0.43
fviz_cluster(res.hc) # scatter plot

Infos
This analysis has been performed using R software (R version 3.3.2)