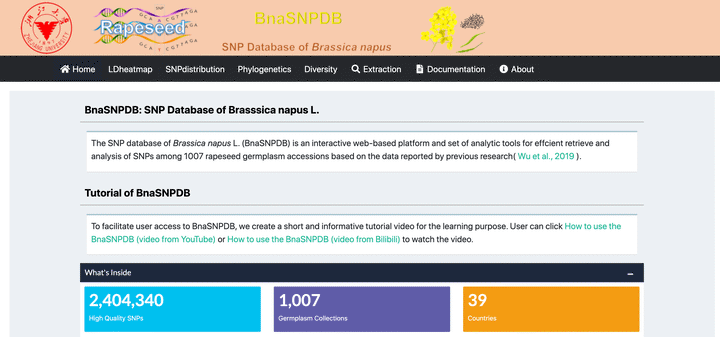
 The home page
The home page
Overview
The SNP database of Brassica napus (BnaSNPDB) is an interactive web portal that provides multiple analysis modules to visualize and explore SNPs among 1007 rapeseed germplasm accessions based on the data reported by previous research ( Wu et al., 2019). The app is deployed at https://bnapus-zju.com/bnasnpdb for online use.
BnaSNPDB is idle until you activate it by accessing the URL. So it may take some time to load for the first time. Once it was activated, BnaSNPDB could be used smoothly and easily.
The portal is built entirely in R and Shiny using the RStudio development environment.
Install
Requirements
- R: https://www.r-project.org/ v4.0.0+
- RStudio: https://rstudio.com/products/rstudio/download
- Shiny Server: https://rstudio.com/products/shiny/download-server (only required for deploying BnaSNPDB on web linux server)
Initialize app
To run the app locally:
- Clone this repository
git clone https://github.com/YTLogos/BnaSNPDB.git
The repository is large so it may need some time to finish it.
-
Open
BnaSNPDB.Rproj -
Install packages. In the RStudio console, run:
# try an http CRAN (Bioc) mirror if https CRAN (Bioc) mirror doesn't work
# First install Bioconductor
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(version = "3.13")
BiocManager::install("shiny",update = F)
BiocManager::install("ggplot2",update = F)
BiocManager::install("stringr",update = F)
BiocManager::install("dplyr",update = F)
BiocManager::install("tidyr",update = F)
BiocManager::install("forcats",update = F)
BiocManager::install("patchwork",update = F)
BiocManager::install("glue",update = F)
BiocManager::install("ggpubr",update = F)
BiocManager::install("writexl",update = F)
BiocManager::install("snpStats"),update = F
BiocManager::install("IRanges",update = F)
BiocManager::install("LDheatmap",update = F)
BiocManager::install("ape",update = F)
BiocManager::install("pegas",update = F)
BiocManager::install("gridExtra",update = F)
BiocManager::install("grid",update = F)
BiocManager::install("ggtree",update = F)
BiocManager::install("shinycssloaders",update = F)
BiocManager::install("shinydashboard",update = F)
BiocManager::install("shinydisconnect",update = F)
BiocManager::install("shinyWidgets",update = F)
BiocManager::install("gggenes",update = F)
BiocManager::install("DT",update = F)
BiocManager::install("shinythemes",update = F)
BiocManager::install("NAM",update = F)
BiocManager::install("adegenet",update = F)
if (require(devtools)) install.packages("devtools")#if not already installed
devtools::install_github("AnalytixWare/ShinySky")
This may take some time to complete - walk away from your computer, rest your eyes, and catch up on those stretching exercises you are meant to be doing :)
- Start tha app by running
shiny::runApp(launch.browser = TRUE)
Deploy BnaSNPDB on web Linux server
- Clone/Upload this repository into /srv/shiny-server
$ cd /srv/shiny-server
git clone https://github.com/YTLogos/BnaSNPDB.git
# Or clone it locally and upload the directory to /srv/shiny-server using scp or other tools
- Configure Shiny Server (/etc/shiny-server/shiny-server.conf)
# Instruct Shiny Server to run applications as the user "shiny"
preserve_logs true;
sanitize_errors false;
run_as shiny;
# Define a server that listens on port 3838
server {
listen 3838;
# Define a location at the base URL
location / {
# Host the directory of Shiny Apps stored in this directory
site_dir /srv/shiny-server;
# Log all Shiny output to files in this directory
log_dir /var/log/shiny-server;
# When a user visits the base URL rather than a particular application,
# an index of the applications available in this directory will be shown.
directory_index on;
}
}
- Change the owner of the BnaSNPDB directory
$ chown -R shiny /srv/shiny-server/BnaSNPDB
- Start Shiny-Server
$ start shiny-server
Now you can access the BnaSNPDB app at http://IPAddressOfYourServer:3838/BnaSNPDB.