rehh:全基因组选择印记扫描

简介
Sabeti在2002年的时候提出了一种叫做Extended Haplotype Homozygosity (EHH)的统计方法,用来扫描全基因组范围内的选择印记。在此基础上又衍生出了iHS、XP-EHH以及Rsb等统计方法。R包rehh实现了这几种统计方法的封装。
应用
rehh支持非常多类型的输入数据,这里我们使用用的较多的数据类型vcf数据,其他的数据类型可以查看
vignette 。
数据输入
该包提供了一些示例数据,这里我们使用的是提供的bta12_cgu.vcf.gz数据
$ zcat bta12_cgu.vcf.gz|less
##fileformat=VCFv4.2
##reference=Btau20070913-freeze
##INFO=<ID=NS,Number=1,Type=Integer,Description="Number of Samples With Data">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT CGU_MN026 CGU_MN027
12 79823 F1200140 T A . PASS NS=140 GT 1|1 0|1 1|1
12 125974 F1200150 C G . PASS NS=140 GT 1|1 1|0 1|0
12 175087 F1200170 G C . PASS NS=140 GT 0|0 1|1 1|1
12 219152 F1200180 A T . PASS NS=140 GT 1|1 1|1 1|1
12 256896 F1200190 T A . PASS NS=140 GT 0|1 1|1 1|1
12 316254 F1200210 C G . PASS NS=140 GT 1|0 0|1 0|0
读取vcf数据之前,先安装R包vcfR
if(!require(vcfR))
BiocManager::install("vcfR")
if(!require(rehh))
install.packages("rehh")
## 数据读取、转换,rehh提供了一个函数data2haplohh
hh <- data2haplohh(hap_file = "bta12_cgu.vcf.gz",
polarize_vcf = FALSE)
* Reading input file(s) *
Scanning file to determine attributes.
File attributes:
meta lines: 4
header_line: 5
variant count: 1424
column count: 149
Meta line 4 read in.
All meta lines processed.
gt matrix initialized.
Character matrix gt created.
Character matrix gt rows: 1424
Character matrix gt cols: 149
skip: 0
nrows: 1424
row_num: 0
Processed variant: 1424
All variants processed
Extracting map information.
Extracting haplotypes.
Number of individuals which are
Haploid Diploid Triploid, ... :
1 2
0 140
* Filtering data *
Discard markers genotyped on less than 100 % of haplotypes.
No marker discarded.
Data consists of 280 haplotypes and 1424 markers.
Number of mono-, bi-, multi-allelic markers:
1 2
27 1397
计算EHHS、可视化
#这里我们选择SNP位点F1205400
res <- calc_ehhs(hh,
mrk = "F1205400",
include_nhaplo = TRUE)
#可视化EHHS
plot(res)
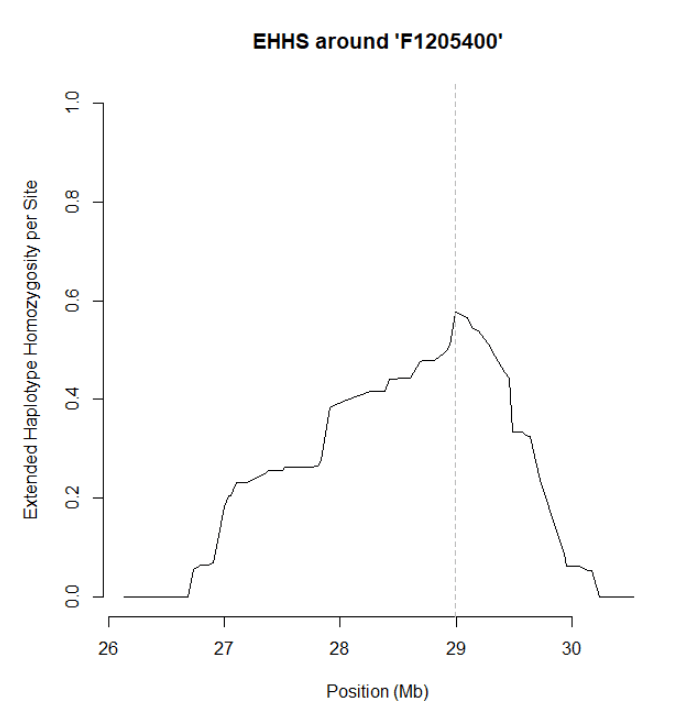
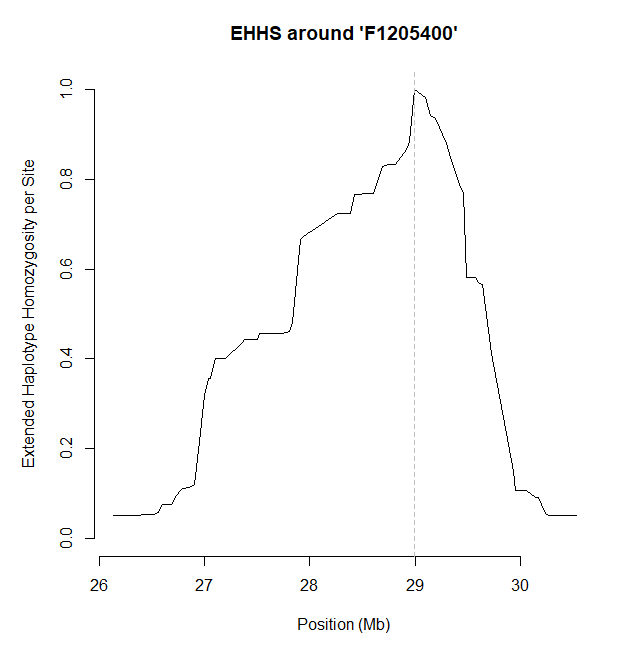
calc_ehhs会计算出EHHS以及nEHHS(un-normalized EHHS),plot默认可视化的是EHHS,也可以绘制nEHHS,此时focal marker也就是我们这里选的F1205400SNP处的EHHS的值会被归一化为1.
plot(res, nehhs = TRUE)
数据实战
下面使用我们自己的数据来分析,前期我们重测序了1000份油菜,分为三类生态型:春型、半冬型、冬型。我们可以分别检测这三个群体某个位点的选择印记。
#首先提取各个群体的SNP,准备需要提取的样品名称,这里提取S、SW、W
$ less S.txt
R4168
R4190
R4205
R4210
R4216
...
#提取SNP
/public/home/jianglix/biosoft/vcftools/bin/vcftools --gzvcf Bna.snp.maf.0.05.int.0.5.vcf.gz --keep ./s --recode --recode-INFO-all -c > pop-S.vcf
/public/home/jianglix/biosoft/vcftools/bin/vcftools --gzvcf Bna.snp.maf.0.05.int.0.5.vcf.gz --keep ./w --recode --recode-INFO-all -c > pop-W.vcf
/public/home/jianglix/biosoft/vcftools/bin/vcftools --gzvcf Bna.snp.maf.0.05.int.0.5.vcf.gz --keep ./sw --recode --recode-INFO-all -c > pop-SW.vcf
#建立tabix索引
~/biosoft/htslib1.9/bin/bgzip < pop-S.vcf > pop-S.vcf.gz
~/biosoft/htslib1.9/bin/tabix -p vcf pop-S.vcf.gz
~/biosoft/htslib1.9/bin/bgzip < pop-SW.vcf > pop-SW.vcf.gz
~/biosoft/htslib1.9/bin/tabix -p vcf pop-SW.vcf.gz
~/biosoft/htslib1.9/bin/bgzip < pop-W.vcf > pop-W.vcf.gz
~/biosoft/htslib1.9/bin/tabix -p vcf pop-W.vcf.gz
##如果SNP缺失过多,可以基因型填补,因为缺失太多会影响后续分析
java -jar -Xmx110G "-Djava.io.tmpdir=/database/tmp/" ~/biosoft/beagle/beagle5.jar gt=pop-S.vcf.gz out=pop-S.imputed
java -jar -Xmx110G "-Djava.io.tmpdir=/database/tmp/" ~/biosoft/beagle/beagle5.jar gt=pop-SW.vcf.gz out=pop-SW.imputed
java -jar -Xmx110G "-Djava.io.tmpdir=/database/tmp/" ~/biosoft/beagle/beagle5.jar gt=pop-W.vcf.gz out=pop-W.imputed
基因选取
我们可以选择某些基因区域来查看此基因在不同的群体(春、冬、半冬)中是否受到选择,这里我们选择开花相关基因FLC.A10 (20022944~20027730)。选取基因上下游50KB进行分析
#bed文件,用于提取SNP
10 19982944 20067730
#SNP提取
~/biosoft/htslib1.9/bin/tabix -R flc.A10.bed -h pop-S-imputed.vcf.gz > pop-S.flc.A10.vcf
~/biosoft/htslib1.9/bin/tabix -R flc.A10.bed -h pop-SW-imputed.vcf.gz > pop-SW.flc.A10.vcf
~/biosoft/htslib1.9/bin/tabix -R flc.A10.bed -h pop-W-imputed.vcf.gz> pop-W.flc.A10.vcf
EHHS分析
一般来说是根据GWAS或者其它选择性清除分析结果来选取focal marker,这里我就随便选个SNP(10_20025969)
library(rehh)
library(tidyverse)
s <- data2haplohh("pop-S.flc.A10.vcf", polarize_vcf = FALSE)
sw <- data2haplohh("pop-SW.flc.A10.vcf", polarize_vcf = FALSE)
w <- data2haplohh("pop-W.flc.A10.vcf", polarize_vcf = FALSE)
snp <- "10_20025969"
res_s <- calc_ehhs(
s,
mrk = snp,
include_nhaplo = TRUE
)
res_sw <- calc_ehhs(
sw,
mrk = snp,
include_nhaplo = TRUE
)
res_w <- calc_ehhs(
w,
mrk = snp,
include_nhaplo = TRUE
)
data_s <- res_s$ehhs %>%
mutate(pos = POSITION / 1000000) %>%
mutate(type = "Spring")
data_sw <- res_sw$ehhs %>%
mutate(pos = POSITION / 1000000) %>%
mutate(type = "Semi-Winter")
data_w <- res_w$ehhs %>%
mutate(pos = POSITION / 1000000) %>%
mutate(type = "Winter")
data_all <- rbind(data_s, data_sw, data_w)
p <- ggplot(data_all, aes(pos,NEHHS,color=type))+
geom_line(size=1.25)+
theme_classic()+
theme(legend.title = element_blank(), legend.position = c(0.75,0.85))+
scale_color_manual(values = c("#22ac38","#6299fd","#63c2cb"))+
labs(x="Chromosome A10 (Mb)", y="EHHS")
print(p)

可以看出差异不大,春冬半冬之间的受选择差异不大,这有可能跟挑选的位点有关。
SessionInfo
devtools::session_info()
- Session info -------------------------------------------------------------------------------------------------------------------------------------------------
setting value
version R version 3.6.1 (2019-07-05)
os Windows 10 x64
system x86_64, mingw32
ui RStudio
language (EN)
collate Chinese (Simplified)_China.936
ctype Chinese (Simplified)_China.936
tz Asia/Taipei
date 2019-08-14
- Packages -----------------------------------------------------------------------------------------------------------------------------------------------------
package * version date lib source
ape 5.3 2019-03-17 [1] CRAN (R 3.6.0)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.0)
backports 1.1.4 2019-04-10 [1] CRAN (R 3.6.0)
broom 0.5.2 2019-04-07 [1] CRAN (R 3.6.0)
callr 3.3.1 2019-07-18 [1] CRAN (R 3.6.1)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.0)
cli 1.1.0 2019-03-19 [1] CRAN (R 3.6.0)
cluster 2.1.0 2019-06-19 [2] CRAN (R 3.6.1)
colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.0)
crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.0)
desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.0)
devtools 2.1.0 2019-07-06 [1] CRAN (R 3.6.1)
digest 0.6.20 2019-07-04 [1] CRAN (R 3.6.1)
dplyr * 0.8.3 2019-07-04 [1] CRAN (R 3.6.1)
fansi 0.4.0 2018-10-05 [1] CRAN (R 3.6.0)
forcats * 0.4.0 2019-02-17 [1] CRAN (R 3.6.0)
fs 1.3.1 2019-05-06 [1] CRAN (R 3.6.0)
gapminder * 0.3.0 2017-10-31 [2] CRAN (R 3.6.0)
generics 0.0.2 2018-11-29 [2] CRAN (R 3.6.0)
ggplot2 * 3.2.1 2019-08-10 [1] CRAN (R 3.6.1)
glue 1.3.1 2019-03-12 [1] CRAN (R 3.6.0)
grkstyle 0.0.1 2019-08-13 [1] Github (gadenbuie/grkstyle@a141d39)
gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.0)
haven 2.1.1 2019-07-04 [1] CRAN (R 3.6.1)
hms 0.5.0 2019-07-09 [1] CRAN (R 3.6.1)
httr 1.4.1 2019-08-05 [1] CRAN (R 3.6.1)
jsonlite 1.6 2018-12-07 [1] CRAN (R 3.6.0)
labeling 0.3 2014-08-23 [1] CRAN (R 3.6.0)
lattice 0.20-38 2018-11-04 [2] CRAN (R 3.6.1)
lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.6.0)
lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.6.0)
magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.0)
MASS 7.3-51.4 2019-03-31 [2] CRAN (R 3.6.1)
Matrix 1.2-17 2019-03-22 [1] CRAN (R 3.6.0)
memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.0)
memuse 4.0-0 2017-11-10 [1] CRAN (R 3.6.0)
mgcv 1.8-28 2019-03-21 [2] CRAN (R 3.6.1)
modelr 0.1.5 2019-08-08 [1] CRAN (R 3.6.1)
munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.0)
nlme 3.1-141 2019-08-01 [2] CRAN (R 3.6.1)
permute 0.9-5 2019-03-12 [1] CRAN (R 3.6.0)
pillar 1.4.2 2019-06-29 [1] CRAN (R 3.6.0)
pinfsc50 1.1.0 2016-12-02 [1] CRAN (R 3.6.0)
pkgbuild 1.0.4 2019-08-05 [1] CRAN (R 3.6.1)
pkgconfig 2.0.2 2018-08-16 [1] CRAN (R 3.6.0)
pkgload 1.0.2 2018-10-29 [2] CRAN (R 3.6.0)
prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.6.0)
processx 3.4.1 2019-07-18 [1] CRAN (R 3.6.1)
ps 1.3.0 2018-12-21 [2] CRAN (R 3.6.0)
purrr * 0.3.2 2019-03-15 [1] CRAN (R 3.6.0)
R6 2.4.0 2019-02-14 [1] CRAN (R 3.6.0)
Rcpp 1.0.2 2019-07-25 [1] CRAN (R 3.6.1)
readr * 1.3.1 2018-12-21 [1] CRAN (R 3.6.0)
readxl 1.3.1 2019-03-13 [1] CRAN (R 3.6.0)
rehh * 3.0.1 2019-07-11 [1] CRAN (R 3.6.1)
rehh.data 1.0.0 2016-11-08 [1] CRAN (R 3.6.0)
remotes 2.1.0 2019-06-24 [2] CRAN (R 3.6.0)
rlang 0.4.0 2019-06-25 [1] CRAN (R 3.6.0)
rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.0)
rstudioapi 0.10 2019-03-19 [1] CRAN (R 3.6.0)
rvest 0.3.4 2019-05-15 [1] CRAN (R 3.6.0)
scales 1.0.0 2018-08-09 [1] CRAN (R 3.6.0)
sessioninfo 1.1.1 2018-11-05 [2] CRAN (R 3.6.0)
stringi 1.4.3 2019-03-12 [1] CRAN (R 3.6.0)
stringr * 1.4.0 2019-02-10 [1] CRAN (R 3.6.0)
styler 1.1.1 2019-05-06 [1] CRAN (R 3.6.0)
testthat 2.2.1 2019-07-25 [1] CRAN (R 3.6.1)
tibble * 2.1.3 2019-06-06 [1] CRAN (R 3.6.0)
tidyr * 0.8.3 2019-03-01 [1] CRAN (R 3.6.0)
tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.6.0)
tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.6.0)
usethis 1.5.1 2019-07-04 [1] CRAN (R 3.6.1)
utf8 1.1.4 2018-05-24 [1] CRAN (R 3.6.0)
vcfR 1.8.0 2018-04-17 [1] CRAN (R 3.6.0)
vctrs 0.2.0 2019-07-05 [1] CRAN (R 3.6.1)
vegan 2.5-5 2019-05-12 [1] CRAN (R 3.6.0)
viridisLite 0.3.0 2018-02-01 [1] CRAN (R 3.6.0)
withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.0)
xml2 1.2.2 2019-08-09 [1] CRAN (R 3.6.1)
zeallot 0.1.0 2018-01-28 [1] CRAN (R 3.6.0)
[1] C:/Tools/R/R_Library
[2] C:/Tools/R-3.6.1/library